Figure 4.
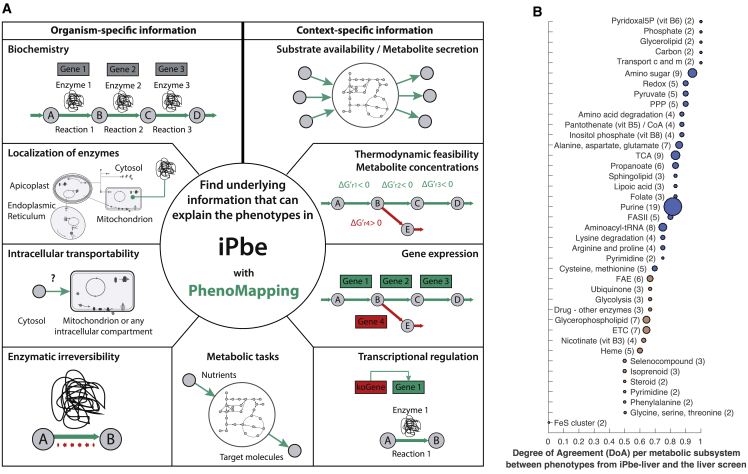
The PhenoMapping Workflow and Degree of Agreement for Metabolic Subsystems in iPbe-Liver with the M-L Screen
(A) Illustration of the PhenoMapping workflow for the integration of organism- and context-specific information into the genome-scale iPbe metabolic models. Context-specific information denotes life-cycle stage-specific processes, such as gene expression, as well as environmentally specific factors, such as substrate availability. Metabolic tasks are at the interface between organism- and context-specific information. The production of molecules, such as amino acids, fatty acids, nucleotides, etc., is required for growth independent of the context, but the ratios in which they are required might change with the growing conditions or life stage. See STAR Methods and Table S4 for a detailed description of iPbe.
(B) Degree of agreement (DoA) between the gene essentiality predictions in iPbe-liver and the experimental phenotypes at the SG-B2 transition. Metabolic subsystems are ranked by level of agreement. Numbers show genes with screen data per subsystem (needs to be >1 for inclusion).