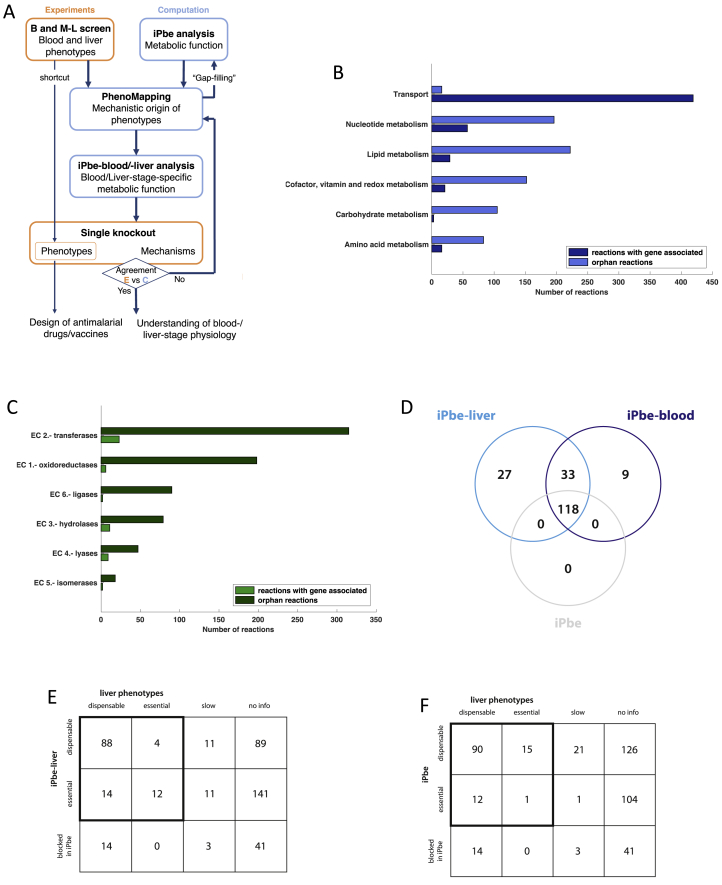
Figure S2.
Combined Experimental and Computational Workflow to Study Blood and Liver Phenotypes and Their Mechanistic Origin using iPbe. Description of the iPbe Model and Essentiality Predictions, Related to Figure 4
(A) Workflow diagram showing how data form the experimental screening platform are integrated to study blood and liver phenotypes and their underlying mechanisms and to develop a more comprehensive metabolic model following the cycle of systems biology. (B) Distribution of metabolic enzymes in iPbe. (C) Metabolic subsystems in iPbe. (D) Relation between genes predicted as essential in iPbe, iPbe-blood and iPbe-liver. (E) Contingency matrix for gene essentiality predictions and the liver stage M-L screen phenotypes compared with iPbe liver. (F) Contingency matrix as for (E) but compared with the general iPbe model.