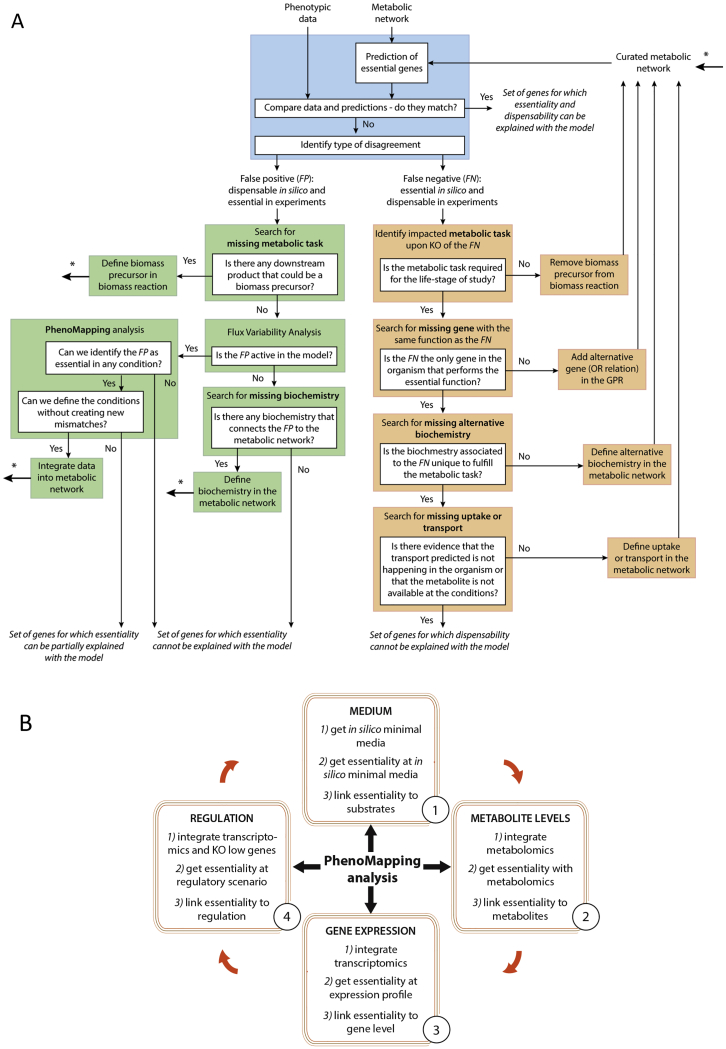
Figure S8.
The PhenoMapping Workflow to Curate Metabolic Models and PhenoMapping Analyses to Identify Context-Specific Conditions Underlying Phenotypes, Related to Figure 4
(A) The PhenoMapping workflow involves three sets of analyses: (i) essentiality studies using the metabolic model and comparison with phenotypic data (blue); (ii) correction of False Negatives (FN, orange); and correction of False Positives (FP, blue). The curation of the metabolic network provides hypotheses on the metabolic function, here essential genes and underlying processes responsible for the essentiality. The classification of genes as fully, partially, or not agreeing with observed phenotypes might change when one modifies the metabolic network (at each iteration through the workflow). GPR denotes the gene-protein-reaction associations in the metabolic model. (B) PhenoMapping analysis to study four context-specific cellular processes or conditions underlying phenotypes: the uptake of substrates, thermodynamic directionality of reactions and transports for a set of context-specific metabolite concentrations, gene expression, and regulation of gene expression. The PhenoMapping analysis is modular: linear arrows from the center of the figure to the boxes of cellular processes indicate an independent PhenoMapping analysis of cellular processes; and curved arrows between cellular processes indicate that the PhenoMapping analysis can be applied cumulatively following the order number suggested.