Figure 1. H3K36 Methylation Is Required for Efficient Pre-mRNA Splicing.
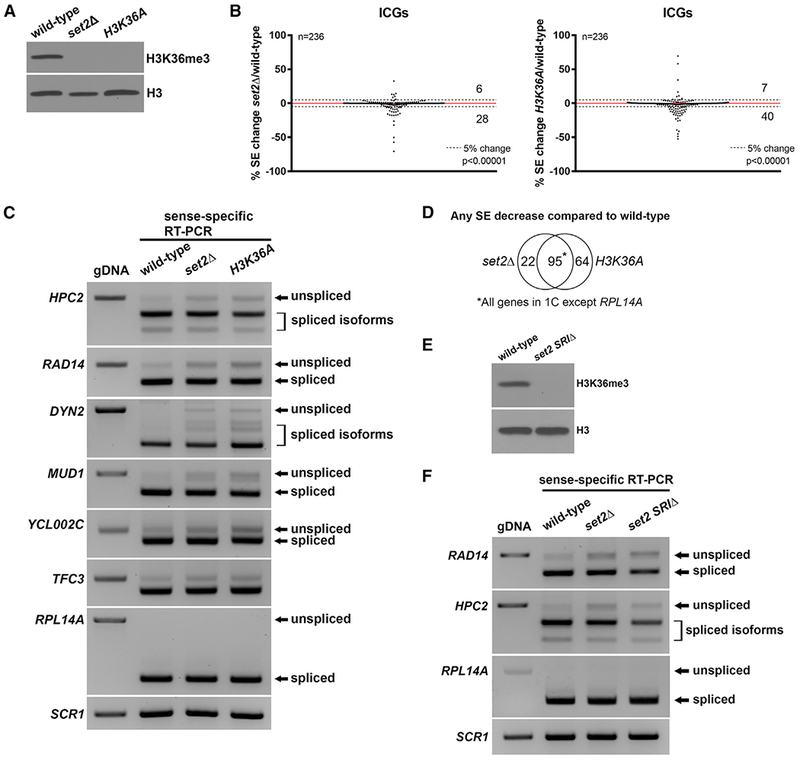
(A) H3K36me3 is absent in the set2Δ and H3K36A mutants. Whole cell extracts from wild-type, set2Δ, and H3K36A cells were subjected to western blotting.
(B) Changes in splicing efficiencies (SEs) of ICGs upon loss of H3K36me represented in a scatterplot. Dashed lines represent a 5% change in SE in the mutant compared to wild-type. Overall splicing decreases in both set2Δ and H3K36A compared with wild-type (chi-square test, p value indicated). Numbers indicate number of ICGs above and below the dashed lines.
(C) RT-PCR validation of splicing changes observed in RNA-seq analysis. ICGs shown display a ≥ 5% decrease in SE in both set2Δ and H3K36A compared with wild-type. RPL14A is a gene that does not show a change in SE. Products were analyzed on a 1.8% agarose gel.
(D) Venn diagram displaying significant overlap of ICGs that display any SE decrease in set2Δ and H3K36A compared with wild-type cells (p < 0.0001, chi-square test).
(E) H3K36me3 is absent in a set2 SRIΔ mutant. Whole cell extracts from wild-type and set2 SRIΔ were subjected to western blotting.
(F) RT-PCR analysis of splicing changes in set2 SRIΔ cells. Products were analyzed on a 1.8% agarose gel. gDNA, genomic DNA. SCR1 is a loading control.
