Figure 2. Eaf3 Is Required for Efficient Pre-mRNA Splicing.
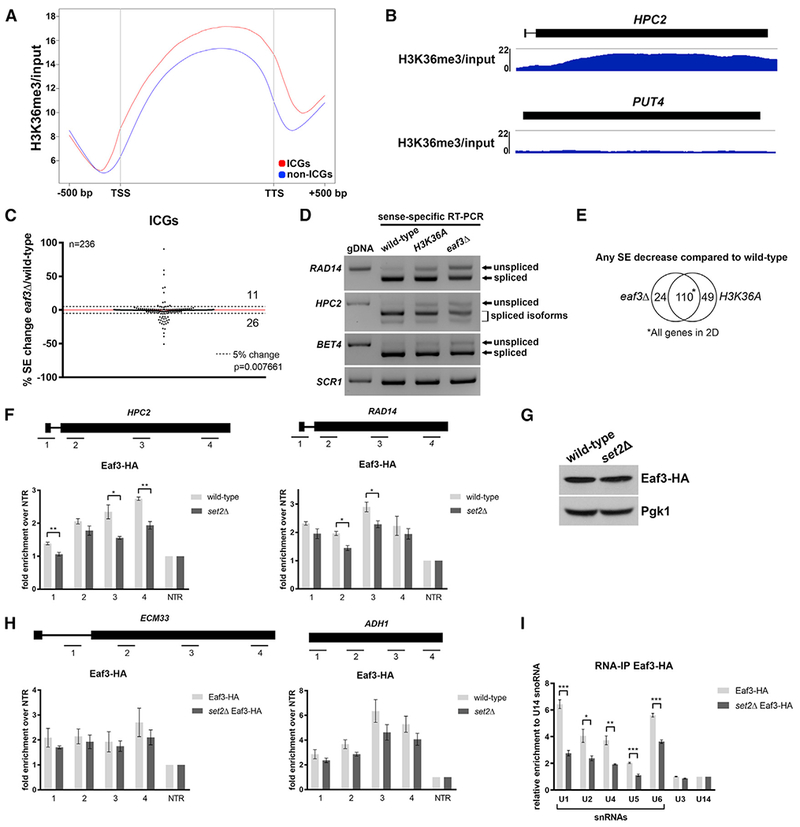
(A) Metagene plot of H3K36me3 ChIP-seq enrichment across the transcribed region and 500 nucleotides (nt) upstream of the transcription start site (TSS) and 500 nt downstream of the transcription termination site (TTS) of ICGs and non-ICGs.
(B) Genome-browser tracks of H3K36me3 ChIP-seq enrichment on ICG HPC2 (RPKM: 7.355; length: 1962 bp) and intronless gene PUT4 (RPKM: 7.658; length: 1884 bp).
(C) Scatterplot of percentage change in splicing efficiency (SE) of all ICGs in eaf3Δ over wild-type. Dashed lines represent a 5% change in SE. Numbers indicate number of ICGs above and below the dashed lines.
(D) RT-PCR validation of splicing changes observed in the eaf3Δ mutant. Products were run on a 1.8% agarose gel. SCR1 is a loading control.
(E) Venn diagram displaying significant overlap of ICGs that display any SE decrease in eaf3Δ and H3K36A compared to wild-type cells (p < 0.0001, chi-square test).
(F) Occupancy of Eaf3-HA on ICGs HPC2 (left) and RAD14 (right) relative to a non-transcribed region (NTR) on chromosome V (Chr. V) in wild-type (light gray bars) and set2Δ cells (dark gray bars).
(G) Western blot analysis of Eaf3-HA protein levels in wild-type and set2Δ cells. Pgk1 is a loading control.
(H) Occupancy of Eaf3-HA on ICG ECM33 relative to a NTR on Chr. V in wild-type and set2Δ cells (left). Occupancy of Eaf3-HA on intronless gene ADH1 relative to a NTR on Chr. V in wild-type and set2Δ cells (right).
(I) Spliceosomal snRNAs co-immunoprecipitated with Eaf3. Bar graph depicting the five spliceosomal snRNAs that are immunoprecipitated with Eaf3 in wild-type and set2Δ cells under non-crosslinking conditions. U3 and U14 are snoRNAs. Bars represent the average of 3 biological replicates. Error bars represent the SEM. p values were determined by 2-tailed unpaired t test. *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001.
