Fig. 4.
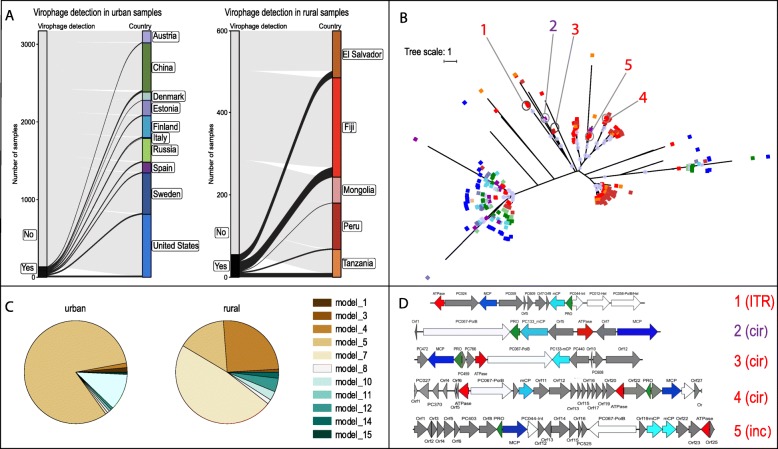
Human gut-associated virophage sequences. a Geographic and lifestyle distribution of the human gut samples queried for the presence of virophages. Gray denotes samples with no hits to any of the MCP models, while black colors correspond to samples with hits to different MCP models from the indicated country. b Unrooted maximum likelihood phylogenetic tree of the 353 MCP sequences detected in the human gut samples. Branch support values > 90% are shown at each node using purple circles. Colored squares at the tip of the branches indicates the country of the sample according to the color code of panel a: “warm colors” (red, brown, orange, amber) or “cold colors” (blues, greens, and purples) represent samples from countries with rural or westernized lifestyles according to sample metadata, respectively. MCP genes found in sequences longer than 10 kb are indicated with numbers 1–5 and colored according to the country where they were detected. c Proportion of the MCP sequences detected by different HMM models (corresponding to different colors as indicated) in westernized and rural lifestyles. d Genetic organization of the 5 gut virophage genomes longer than 10 kb. The four core genes were colored as follows: red denotes ATPase, dark blue MCP, light blue mCP, and green PRO. Other common genes (in white) or unknown genes (in gray) are also displayed and their protein cluster (PC) or annotation indicated when possible (Int, integrase; Hel, helicase; PolB, polymerase B). Numbers 1–5 and their colors correspond to the same numbers and sample colors shown in panel b. 1, SRS475626|k119_215568 (17,831 bp; clade 8); 2, ERS396424|k79_177141 (12,062 bp; clade 11); 3, SRS476271|k119_132073 (17,103; clade 12); 4, SRS476076|k119_199462 (34,763 bp; clade 10); 5, SRS476192|k119_38656 (31,481 bp; clade 12). The circularity (cir) or the incompleteness of the genome (inc), as well as the presence of an inverted terminal repeat (ITR), are indicated next to the number