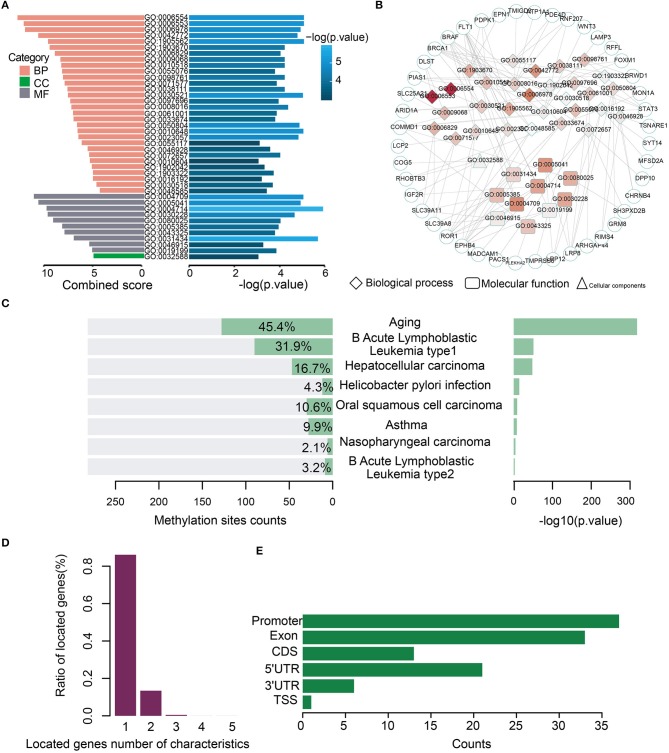
Figure 3.
Biological processes and phenotypic traits of age-related methylation sites for CancerClock. (A) The barplot shows the combined score and –log (p-values) for enrichment GO terms of 282 extracted methylation sites. Pink, green, and gray colors represent biological process (BP), molecular function (MF), and cellular component (CC), respectively. (B) Network shows the interactions between GO terms and genes. The color of node in network indicates enrichment strength, and the three different shapes represent different biological types of GO terms. The circle represents the genes. (C) The bar chart shows the enrichment counts and significance P-values of each trait from EWAS atlas analysis. (D) The corresponding relationship between methylation site and the genes in which it is located. The relationship is usually one-to-one or one-to-many. (E) The distribution of 282 features of CancerClock model in genome position.