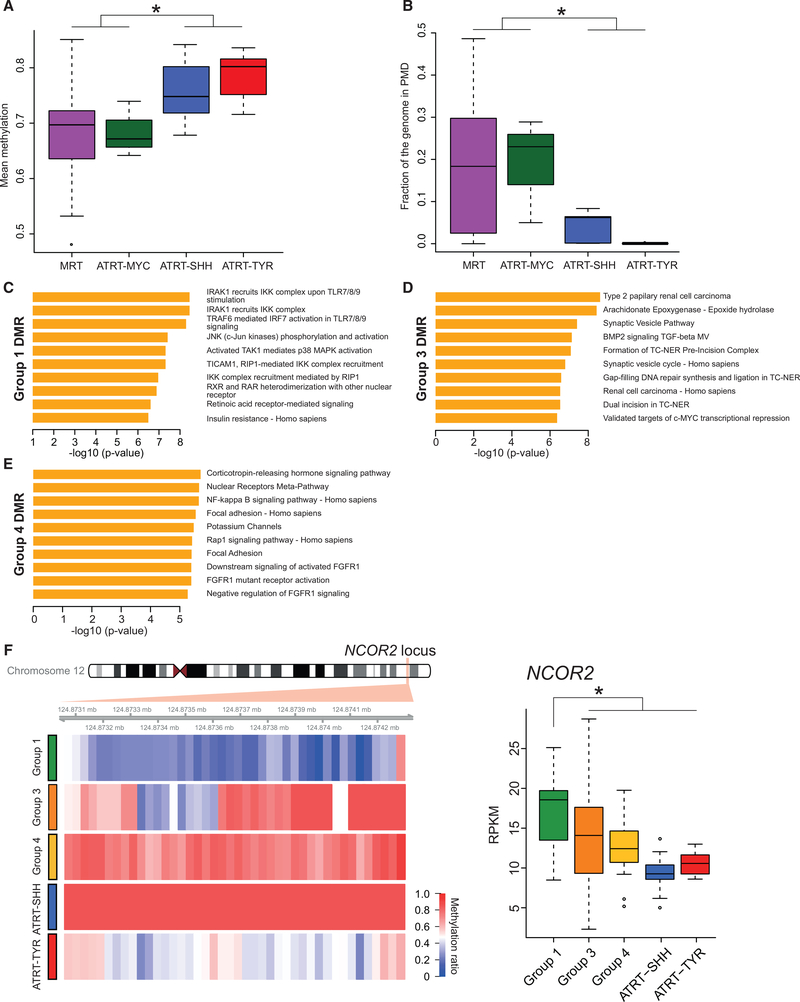
Figure 3. ATRT-MYC and MRT Exhibit Similar DNA Methylation Profiles Distinct from ATRT-SHH and -TYR.
(A) Boxplot shows the distribution of mean genome-wide DNA methylation levels based on WGBS data. MRT (n = 69 cases) and ATRT-MYC (n = 3 cases) showed significant hypomethylation compared to ATRT-SHH (n = 7 cases) and -TYR (n = 7 cases; *Wilcoxon p value < 0.05).
(B) Boxplot displays the distribution of fractions of the genome covered by PMDs in MRT and ATRT-MYC, which exhibited significantly more abundant PMDs compared to ATRT-SHH and -TYR (*Wilcoxon p value < 0.05).
(C–E) Gene set enrichment of DMRs that are specific for Groups 1 (C), 3 (D), and 4 (E). The x axes indicate the statistical significance of the enrichment test. (F) Heatmap (left) shows average CpG methylation levels at the NCOR2 locus in Group-1-specific DMRs (red = 100%; blue = 0% methylation). Boxplot (right) shows significantly increased NCOR2 expression levels in Group 1 compared to other RT subgroups (*Wilcoxon p value < 0.05).
See also Figure S4 and Table S3.