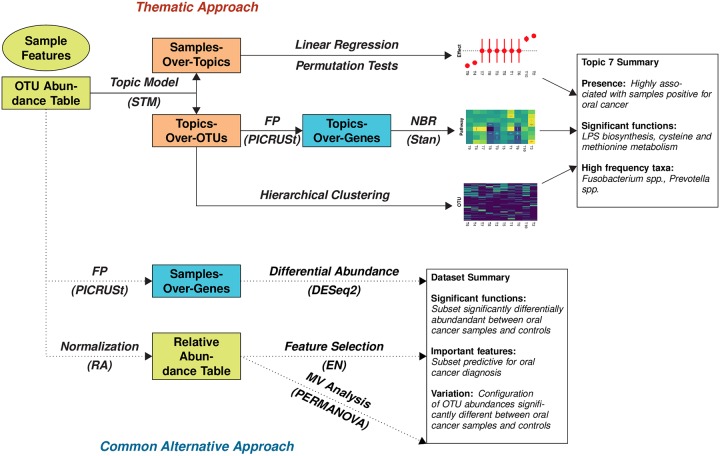
Fig 1.
(Thematic Approach) Given a 16S rRNA gene abundance table, a topic model is used to uncover the thematic structure of the data in the form of two latent distributions: The samples-over-topics frequencies and the topics-over-OTUs frequencies. The samples-over-topics frequencies are regressed against sample features of interest to identify the strength of a topic-covariate relationship to rank topics (top). The topics-over-OTUs frequencies are used in a gene function prediction (FP) algorithm to predict gene content. Important functional categories are identified via a fully Bayesian multilevel negative binomial (NBR) regression model (middle). The topics-over-OTUs distribution is hierarchically clustered to infer relationships between clusters of co-occurring OTUs and topics (bottom). The result is the ability to identify key topics that associate clusters of bacteria and their associated functional content to sample information of interest. (Alternative Approach). A common alternative approach currently used in the literature involves independently (1) characterizing the taxonomic configuration and (2) predicting the functional configuration of the OTU abundance table. Gene function prediction is performed on the full OTU abundance table, followed by a differential abundance analysis to infer differences in specific genes between sample features of interest (top). The OTU table is normalized to overcome library size inconsistencies and then analyzed via two methods: (1) an elastic net (EN) to find sparse sets of OTUs that are predictive for the sample feature of interest (middle) and (2) a multivariate (MV) analysis to identify relationships between beta diversity and the sample feature of interest (bottom). The result are three analyses that summarize the entire OTU relative abundance table, unlike the thematic approach, which characterizes co-occurring sets of OTUs (configurations) in three ways.