Fig. 1. SOX TFs are enriched in the stomach as the chromatin landscape acquires accessibility during development.
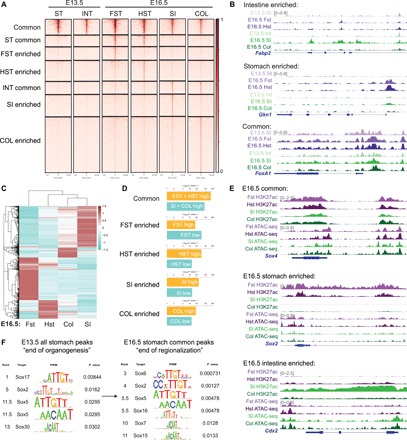
(A) ATAC-seq profiles of E13.5 stomach and intestine compared to E16.5 forestomach, hindstomach, intestine, and colon. Region-enriched peaks are determined on the basis of E16.5 profiles, and their corresponding accessibility is analyzed in the E13.5 tissues. (B) Representative examples of ATAC-seq tracks show acquired chromatin accessibility in E16.5 stomach-specific (Gkn1) and intestinal-specific (Fabp2) loci as the GI organs become regionalized, while common, endodermal loci such as FoxA1 are accessible throughout gut development. (C) Unsupervised hierarchical clustering of E16.5 forestomach, hindstomach, small intestine, and colon transcriptomes based on the top 2000 most variable genes. (D) Binding and Expression Target Analysis (BETA) comparing association of region-specific ATAC-seq peaks to genes differentially regulated in each region at E16.5. (E) Representative examples of H3K27ac ChIP-seq and ATAC-seq peaks of common (Sox4), stomach-specific (Sox2), and intestinal-specific (Cdx2) active DNA regulatory elements such as promoters and enhancers are shown. (F) TF motif analysis of all E13.5 stomach and E16.5 stomach common ATAC-seq peaks shows enrichment of SOX TF motif. Rank indicates the ranking of the motif enrichment in all 637 mouse TF motifs. For ATAC-seq (n = 1 per group), six to eight embryonic guts were pooled. For H3K27ac ChIP-seq (n = 1 per group), 25 to 30 embryonic guts were pooled. For RNA-seq (n = 2 per group), 13 to 15 embryonic guts were pooled. ST, stomach; INT, intestine; FST, forestomach; HST, hindstomach; SI, small intestine; COL, colon.
