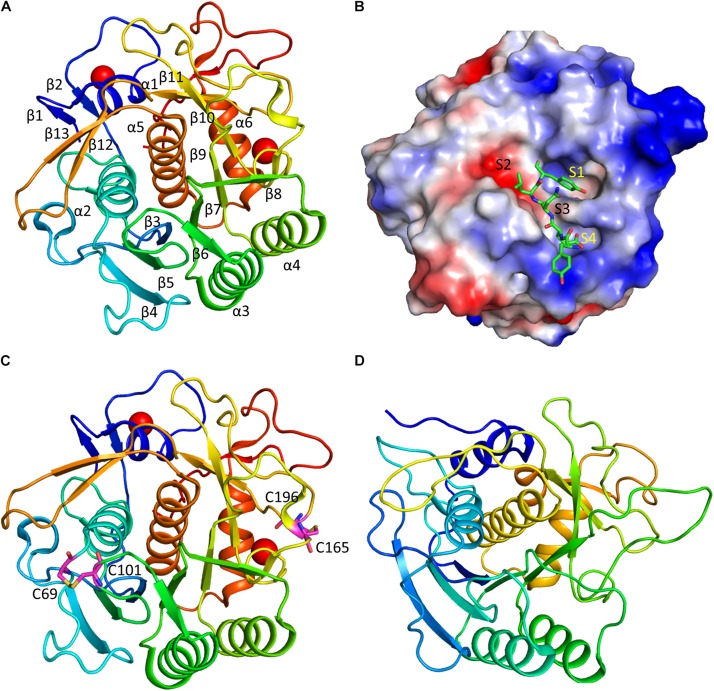
FIGURE 3.
Structure and substrate binding of keratinases. (A) Crystal structure of rMtaKer. The structure of rMtaKer (PDB id 5WSL) is shown. The metal ions are shown as spheres. (B) Surface charge representation of rMtaKer. The substrate binding to the active site are shown as sticks. (C) The residues critical for disulfide bond formation are shown. (D) Crystals structure of other keratinase. The crystal structure of a bacterial keratinase (PDB id 1CSE) is shown. No cysteine residue is present in this enzyme.