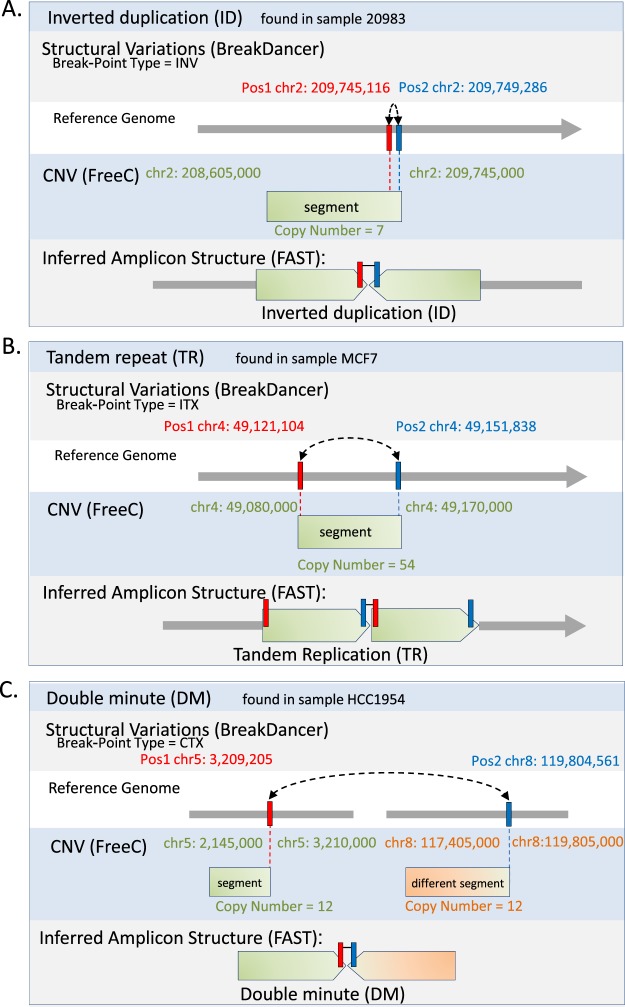
Figure 6.
Identification of amplicon structure (AS) using copy number variation (CNV) and structural variation (SV). Following low coverage whole genome sequencing of tumor DNA we detected and analyzed structural variation (SV) using BreakDancer and enumerated copy number (CN) using Control-FREEC. The data was integrated and further analyzed using FindAmpliconSTructure: FAST, to identify regions with genomic amplification and infer amplicon structure (AS). (A) Identification of inverted duplication (ID) based amplification: A break-point of type Inversion (INV) was found by BreakDancer between Pos1 and Pos2 (chr2: 209,745,116 and chr2: 209,749,286). A segment (chr2:208,605,000-209,745,000 in 7 copies) was found by FreeC, the right edge of which coincides with Pos1 and Pos2. This combination delineates an amplicon structure of Inverted duplication (ID), in which the segment is repeated head-to-head. (B) Identification of tandem repeat (TR) based amplification: A break-point of type Internal-Translocation (ITX) was found by BreakDancer between Pos1 and Pos2 (chr4: 49,121,104 and chr4: 49,151,838). A segment (chr4: 49,080,000-49,170,000 in 54 copies) was found by FreeC, coinciding with Pos1 and Pos2, as shown. This combination delineates an amplicon structure of Tandem Repeat (TR), in which the segment is repeated head-to-tail. (C) Identification of double minute (DM) based amplification: A break-point of type inter-chromosomal translocation (CTX) was found by BreakDancer between Pos1 and Pos2 (chr5: 3,209,205 and chr8: 119,804,561). Two segments were found by FreeC (chr5: 2,145,000-3,210,000 in 12 copies, and chr8: 117,405,000-119,805,000 in 12 copies), one edge of each segment coincides with one of the break-points’ positions. This combination delineates an amplicon structure of Double Minute (DM), in which segments from different chromosomes are linked. The following link to a session in UCSC Browser53 shows the CN and SV tracks of samples.