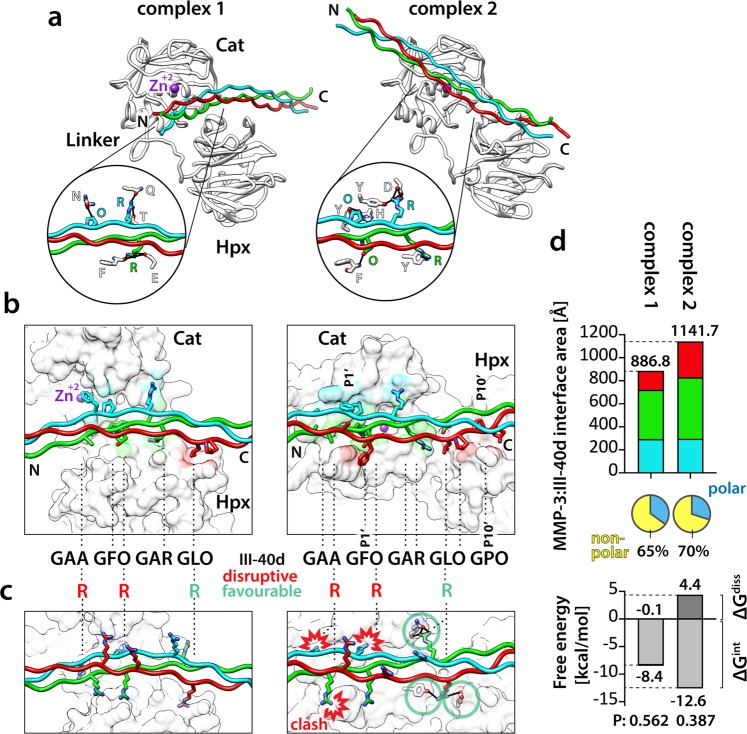
Figure 3.
(a) Two modelled frontal-binding MMP-3:III-40d complexes shown as ribbons in the standard MMP-3 orientation. MMP-3 chain, white; Zn2+, magenta. THP chains: leading, blue; middle, green; trailing, red. Circles, close-up views of MMP-3:III-40d hydrogen bond networks (black lines), with the bonding residues represented as sticks coloured by heteroatom: N, navy blue; O, red, and labelled with 1-letter codes. (b) Close-up view of the modelled MMP-3:III-40d interfaces. The THP is represented as in (a), with side chains containing at least one atom located within 4 Å of the MMP-3 shown as sticks coloured by heteroatom: N, navy blue; O, red. MMP-3 is shown as solvent-accessible surface (UCSF Chimera75) coloured according to the interfacing THP chains (4 Å distance). IDs of the staggered residues in the III-40d THP are roughly indicated (dashed lines). The P1’ and P10’ sites (nomenclature37) are indicated for complex 2. In each complex, all three chains of the THP make contacts with MMP-3. (c) Theoretical analysis of the effects of III-40d residue substitutions on MMP-3 binding. MMP-3 is shown as white and transparent SAS and III-40d is shown as in (a), with modelled Arg side chains and hydrogen-bonded MMP-3 residues represented with sticks coloured by heteroatom: N, navy blue; O, red. Arg at the position Y of the Triplets −1 and 0 causes severe clashes with MMP-3 only in complex 2. (d) Top, NACCESS36 computation of the modelled MMP-3:III-40d interface areas, colour-coded as in (b). Pie charts indicate the polar and nonpolar fractions of the total interface area for each modelled complex. Bottom, PDBePISA computation of the MMP-3:III-40d binding energies in each modelled complex. ΔGint, estimated solvation free energy gain upon complex formation, excluding the effect of satisfied hydrogen bonds and salt bridges across the interfaces. ΔGint < 0 corresponds to hydrophobic interfaces, or positive protein affinity. The ΔGint P-value indicates interface specificity: P = 0.5, not specific; P > 0.5, likely an artefact; P < 0.5, likely interaction-specific. ΔGdiss, estimated free energy needed to dissociate the assembly; interfaces with ΔGdiss > 0 are thermodynamically stable.