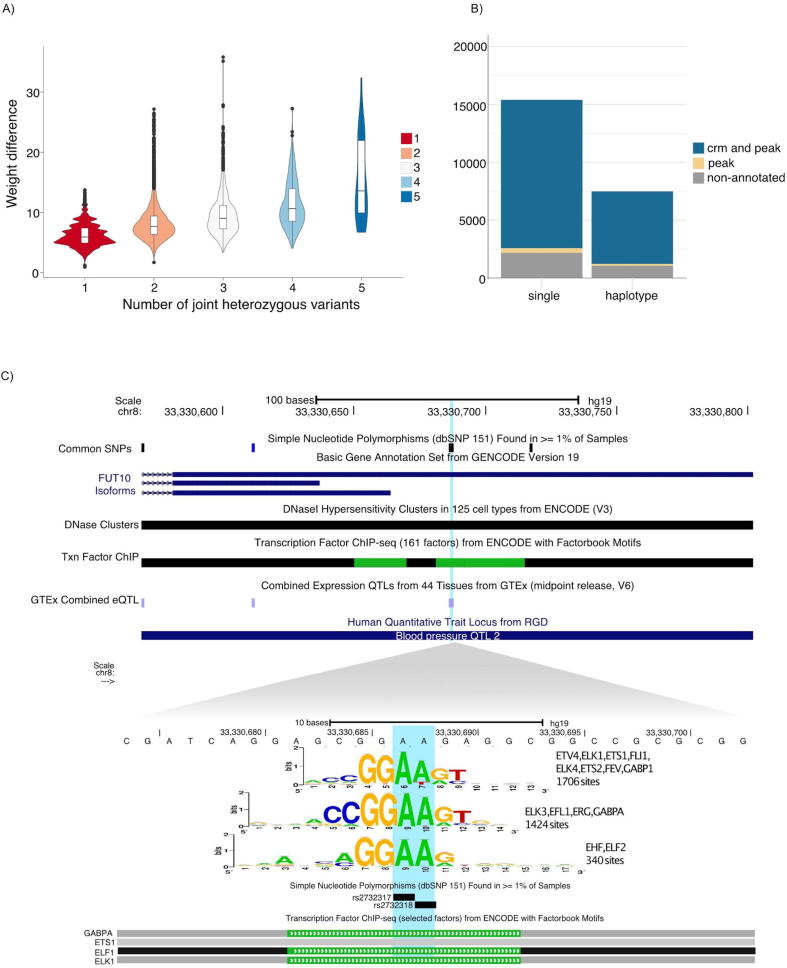
Fig. 3.
Haplotype analysis in high-quality human genomes. A) The number of heterozygous variants (X-axis) within the same putative binding site tend to have a greater impact on the TF binding probability. This is expected as the increase of weight difference observed on the violin plot corresponds to the expected cumulated impact of variations affecting different positions of the same binding site. B) Number of predicted disrupted Transcription Factor Binding Sites (TFBSs) with Cis-Regulatory Modules (CRMs) and TF ChIP-seq peak annotation (blue), with only peak annotation (yellow), and non-annotated predictions (grey). C) University of California Santa Cruz (UCSC) browser [48] screen shot, showing a locus encompassing two SNPs that compose an heterozygous haplotype in one of the Northern Europeans from Utah (CEU) individuals. The figure shows the reference genome haplotype. The variants are located in the FUT10 promoter (top). variation-scan predicts an effect in three motifs that represent binding sites for GABPA, ETS1 and ELF2, factors that have been proven to have binding sites in this region by the ENCODE project. The variant rs2732317 has been associated with effects in gene expression by the GTEx project. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)