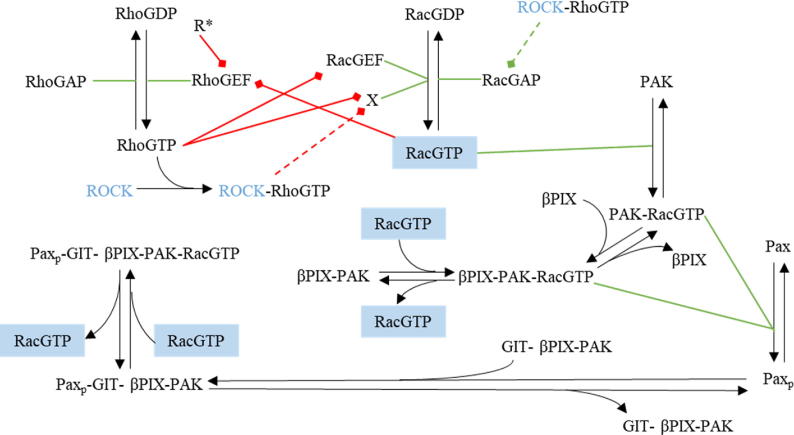
Fig. 1.
Diagram illustrating the interaction between the different proteins involved in cellular motility. Chemical reactions are represented by black arrows, green lines indicate catalysis of a reaction by a specific protein, and red arrows signify inhibition of a protein’s catalytic activity. The dashed lines show plausible mechanisms by which our experimental manipulations could have influenced the biochemical pathways. ROCK is written in blue text to emphasize that it was the target in the experiments. Briefly, the activation/inactivation of the two members of the Rho family of GTPases, Rac and Rho, are regulated by Rac-GEF/Rac-GAP and Rho-GEF/Rho-GAP, respectively, while ROCK activates Rac-GAP. Rac and Rho also mutually inhibit each other’s activation through inhibiting their respective GEFs. Activated Rac activates PAK which in turn induces paxillin phosphorylation at Serine 273 (S273). Phosphorylated paxillin then binds to the protein complex GIT-βPIX-PAK, where βPIX is a Rac-GEF (i.e., can induce Rac activation). Thus, according to this diagram, PAK and βPIX act upstream and downstream of paxillin phosphorylation and Rac activation, respectively. The RacGTP is in blue boxes to indicate that active Rac comes from the same protein pool. Finally, X = [βPIX], [βPIX-PAK], [βPIX-PAK-RacGTP], [GIT-βPIX-PAK], [Paxp-GIT-βPIX-PAK], [Paxp-GIT-βPIX-PAK-RacGTP] (i.e. the set of all protein complexes containing βPIX), whereas (i.e., the set of protein complexes containing RacGTP). (For interpretation of the references to the colours in this figure legend, the reader is referred to the web version of this article.)