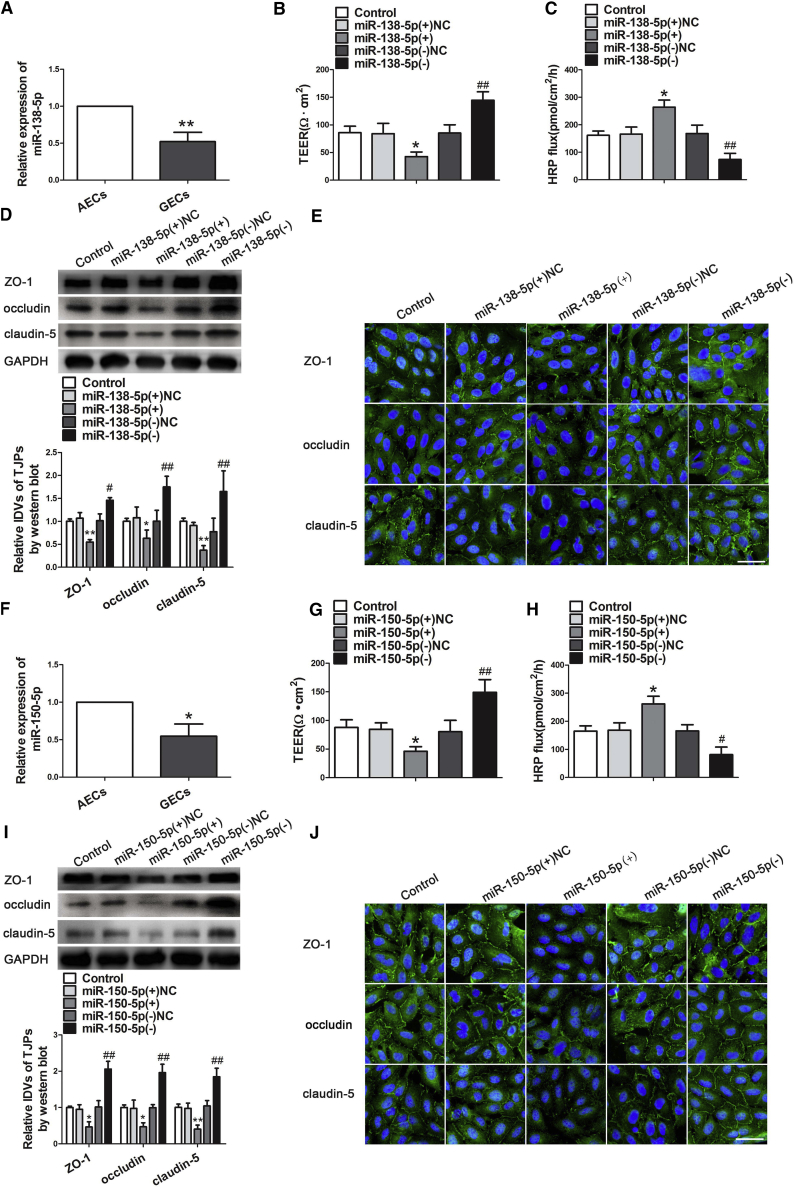
Figure 2.
miR-138-5p and miR-150-5p Regulate BTB Permeability and the Expression of Tight Junction-Related Proteins in GECs
(A) Relative expression levels of miR-138-5p were detected by quantitative real-time PCR in AECs or GECs. Data represent mean ± SD (n = 3, each). **p < 0.01 versus AECs group. (B) TEER values of BTB were detected after overexpression and inhibition of miR-138-5p in GECs. (C) HRP flux assay was performed to detect BTB permeability. Data represent mean ± SD (n = 5, each). *p < 0.05 versus miR-138-5p(+)NC; ##p < 0.01 versus miR-138-5p(−)NC. (D) Western blot analysis of ZO-1, occludin, and claudin-5 in GECs. Data represent mean ± SD (n = 3, each). *p < 0.05 and **p < 0.01 versus miR-138-5p(+)NC; #p < 0.05 and ##p < 0.01 versus miR-138-5p(−)NC. (E) Immunofluorescence localization of ZO-1, occludin, and claudin-5 in GECs. Nuclei are labeled with DAPI. Images are representative of three independent experiments. Scale bar, 50 μm. (F) Relative expression levels of miR-150-5p were detected by quantitative real-time PCR in AECs or GECs. Data represent mean ± SD (n = 3, each). *p < 0.05 versus AECs group. (G) TEER values were detected on BTB integrity, after overexpression and inhibition of miR-150-5p in GECs. (H) HRP flux assay was performed to detect BTB permeability. Data represent mean ± SD (n = 5, each). *p < 0.05 versus miR-150-5p(+)NC; #p < 0.05 and ##p < 0.01 versus miR-150-5p(−)NC. (I) Western blot analysis expression of ZO-1, occludin, and claudin-5 after transfection of miR-150-5p mimics and inhibitor in GECs. Data represent mean ± SD (n = 3, each). *p < 0.05 and **p < 0.01 versus miR-150-5p(+)NC; ##p < 0.01 versus miR-150-5p(−)NC. (J) Immunofluorescence localization of ZO-1, occludin, and claudin-5 in GECs. Nuclei are labeled with DAPI. Images are representative of three independent experiments. Scale bar, 50 μm.