Fig. 1.
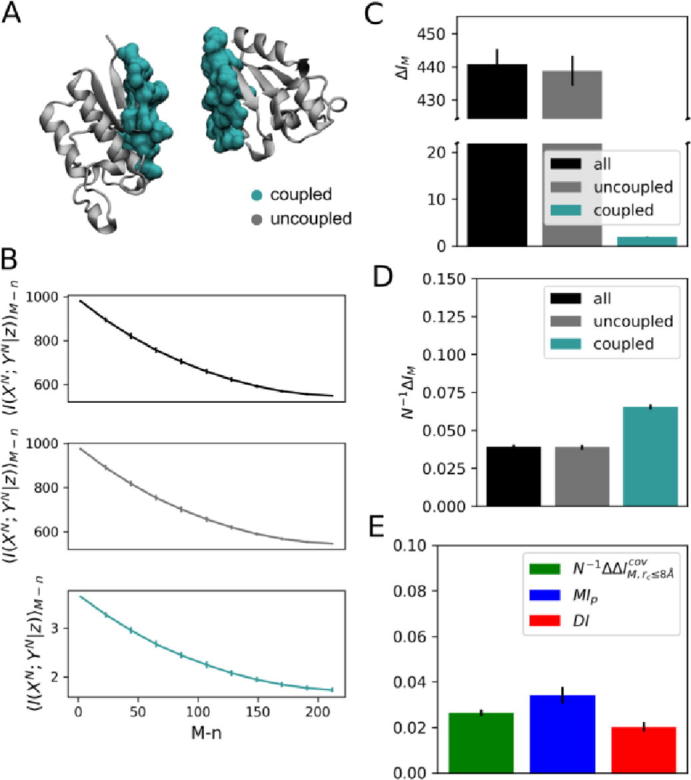
Informational analysis of protein complex TusBCD, chains B and C. (A) Three-dimensional representation of stochastic variables XN and YN as defined from physically coupled amino acids at short-range cutoff distances rc ≤ 8.0 Å (turquoise) and physically uncoupled amino-acids at long-range cutoff distances rc > 8.0 Å (gray). Calculation of rc involved Cβ-Cβ atomic separation distances. (B) Conditional mutual information <I(XN; YN|z)>M−n as a function of the number M − n of randomly paired proteins in the reference (native) MSA, for 0 ≤ n ≤ M. < I(XN; YN|z)>M−n are expectation values estimated from a generated ensemble of 500 MSA models. Mutual information of fully “scrambled” models featuring M unpaired sequences is similar to that calculated from randomized sequence alignments generated by aleatory swapping of lines within columns. (C) Mutual information gap ΔIM between reference and 100 fully “scrambled” models featuring M unpaired sequences. (D) Per-contact mutual information gap N−1ΔIM,rc. (E) Mutual information decomposition according to Eq. (11) and comparison with functional mutual information (MIp,rc≤8Å) and direct information (DIrc≤8Å). In B, C, D and E error bars correspond to standard deviations.
