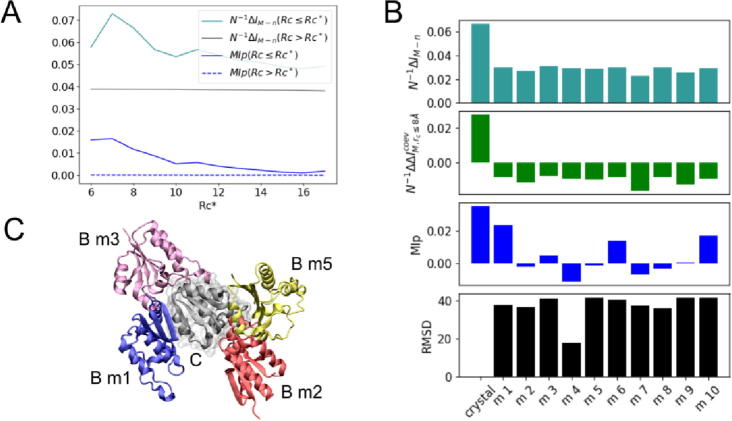
Fig. 3.
Dependence with contact definition rc* and docking decoys. (A) Per-contact mutual information gap N−1ΔIM,rc and mutual information subtracted from structural-functional relationships MIp,rc at various rc*. (B) Per-contact mutual information gap N−1ΔIM,rc (turquoise), information content resulting from coevolution alone N−1ΔΔICovM,rc (green) and mutual information subtracted from structural or functional relationships MIp,rc (blue) at alternative interfaces generated by docking – only physically coupled amino acids as defined for rc ≤ 8.0 Å were included in the calculations. Black bars represent the root-mean-square deviation (RMSD in Ȧ units) between the native bound structure and docking decoys as generated by GRAMM-X [30]. Docking solutions were selected following a stability binding-energy criterium according to the scoring function of GRAMM – all docking decoys considered in the study are low-energy configurations despite large RMSD values relative to the native structure. (C) Illustration of four docking decoys of chain B in the protein complex TusBCD (chain C is shown in gray). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)