Abstract
Papillary thyroid carcinoma (PTC) is the most common malignant tumor of endocrine systems. Chromosomal instability (CIN) is crucial to the clinical prognoses of tumor patients. DNA methylation plays an important role in the regulation of gene expression and CIN. Based on PTC samples from The Cancer Genome Atlas database, we used multiple regression analyses to identify methylation patterns of CpG sites with the strongest correlation with gene expression. A total of 4,997 genes were obtained through combining the CpG sites, which were represented as featured DNA methylation patterns. In order to identify CIN-related epigenetic markers of PTC survival, we developed a method to characterize CIN based on DNA methylation patterns of genes using the Student’s t statistics. We found that 1,239 genes were highly associated with CIN. With the use of the log-rank test, univariate Cox regression analyses, and the Kaplan-Meier method, DNA methylation patterns of UBAC2 and ELOVL2, highly correlated with CIN, provided potential prognostic values for PTC. The higher these two genes, risk scores were correlated with worse PTC patient prognoses. Moreover, the ELOVL2 risk score was significantly different in the four stages of PTC, suggesting that it was related to the progress of PTC. The DNA methylation pattern associated with CIN may therefore be a good predictor of PTC survival.
Keywords: papillary thyroid carcinoma, chromosomal instability, DNA methylation, gene expression, prognosis
Introduction
Papillary thyroid carcinoma (PTC) is derived from the thyroid follicular epithelium. PTC is the most common type of endocrine cancer, and its incidence has increased rapidly over the past several decades.1 It accounts for 85% of thyroid cancer, 60% of adult thyroid cancer,2 and 100% of child thyroid cancer.3 The vast majority of patients are diagnosed with differentiated thyroid carcinoma, especially with PTC.4 This causes difficulty in planning the therapy, because some patients are overtreated, whereas in other patients, the same therapy does not result in the eradication of the neoplastic foci and inhibition of the natural course of the disease. PTCs are usually curable with a 5-year survival of over 95%;5 however, occasionally, they dedifferentiate into more aggressive and lethal thyroid cancers.6 For this reason, it is important to identify effective prognostic markers to evaluate the prognoses of PTC patients.
Commonly used prognostic markers presently include proteins, microRNAs (miRNAs), mRNAs, and DNA methylations. Ma and Yu7 suggested that TBL1XR1 overexpression was an unfavorable prognostic factor for epithelial ovarian cancer, and Wang et al.8 suggested that DHX32 overexpression was an unfavorable prognostic biomarker for breast cancer. The signature of chromosomal instability (CIN), inferred from gene-expression levels, can predict clinical outcomes in multiple human cancers.9 CIN describes a dynamic state in which cells continuously gain or lose whole chromosomes or parts of chromosomes at an elevated rate and is therefore a principal mediator of aneuploidy and intra-tumor heterogeneity.10, 11, 12, 13 Because aneuploidy is a consequence of CIN, genes with expression levels are consistently associated with aneuploidy, so gene-expression signatures provide a means to estimate levels of CIN.9 Carter et al.9 developed a computational method to characterize CIN based on gene-expression levels using the Student’s t statistics. They mapped the genes to chromosomal sub-bands, with CIN describing the net deviation in expression of genes contained in each chromosomal region relative to the remainder of the sampled transcriptome. Patients with a higher CIN score had worse clinical prognoses. They suggested that gene-expression signatures that had high correlations with CIN could therefore predict the clinical prognoses of tumor patients.
Genomic DNA hypomethylation is another important factor associated with CIN.14, 15, 16, 17 Methylation of the carbon-5 position of cytosine, mostly in the context of CpG dinucleotides, is the main epigenetic modification of DNA and is essential for a properly functioning genome, including maintenance of chromosome stability and transcriptional repression.18, 19, 20, 21 Recently, DNA methylation biomarkers for the diagnoses, molecular typing, and prognoses of cancers were identified.22, 23, 24 Lu et al.22 suggested that hypermethylation of hMLH1 in PTC was significantly correlated with age, size, and the number of primary lesions, local invasion, T stage, and lymph node metastases. Shou et al.23 reported that aberrant methylation of the RASSF1A promoter was more frequently detected in thyroid cancer than in noncancerous controls. Wang et al.24 reported that hypermethylation of RUNX3 significantly increased the risk of PTC recurrence by using appropriate site-specific cut-off values.
Genomic DNA hypomethylation has been associated with increased CIN, which plays a central role in tumorigenesis.14, 15, 16, 17 Kawano et al.14 suggested that whole genome hypomethylation initiated carcinogenesis of esophageal squamous cells through CIN. Nishida et al.15 concluded that DNA hypomethylation is an important cause of CIN in the earliest phase of human hepatocellular carcinoma, especially in the background of noncirrhotic livers. Rodriguez et al.16 reported that CIN was correlated with genome-wide DNA demethylation in human primary colorectal cancers, and Suzuki et al.17 reported that global DNA demethylation in gastrointestinal cancer was correlated with increased genomic damage. However, few reports have shown that hypermethylation is associated with CIN.
Gene-expression levels can be affected by a number of factors, including the environment, gene mutations, and DNA methylations.25, 26, 27, 28 Hypermethylated promoters lead to an “off” state of expression, whereas less methylation may lead to an “on” state.19 Methylation is an acquired epigenetic phenomenon but can be faithfully reproduced in the progeny of affected cells, and the methylation will then be propagated during clonal selection during the development of tumors.29 DNA methylations are therefore more stable than gene expressions. Although several methylation biomarkers have been identified to predict cancer survival, they are usually limited to average methylation levels of several genes based on experimental data. However, there is a weak correlation between the average DNA methylation levels of gene promoters and the levels of gene expression.30 This report prompted us to speculate that methylated CpGs might not have equivalent regulatory effects on gene expression, which results in the maximum regulatory effect of DNA methylation on gene expression.31 We then identified the DNA methylation patterns that had high correlations with CIN as prognostic markers of PTC.
In the following study, based on The Cancer Genome Atlas (TCGA) database, we identified differentially methylated CpG sites between PTC and normal samples. Multiple regression analyses were then used to obtain the methylation patterns of CpGs with the highest correlations with gene expression. We obtained specified genes by combining CpG sites, which were represented as specific DNA methylation patterns. In order to identify CIN-related epigenetic markers of PTC survival, a method was developed to characterize CIN based on the DNA methylation patterns of genes using the Student’s t statistics. Pearson’s correlation coefficient (PCC) was used to evaluate the correlations between DNA methylation patterns and the CIN of each gene. With the use of PCC and a permutation test, we verified that the featured genes were highly associated with CIN. With the use of the log-rank test, univariate Cox regression analyses, and the Kaplan-Meier method, we conducted prognostic analyses. The DNA methylation patterns of UBAC2 and ELOVL2 that had high correlations with CIN provided good prognostic values for PTC. Moreover, UBAC2 and ELOVL2 were hypomethylation phenotypes. The DNA methylation patterns associated with CIN may therefore be a good predictor of PTC survival.
Results
Identifying Differentially Methylated CpGs Associated with Gene Expression
With the use of the Illumina Infinium HumanMethylation450 BeadChip assay (Illumina, San Diego, CA), raw data (level 3 data), raw UNC RNAseqV2 level 3 expression data, and the clinical prognostic information for PTC were collected from TCGA. The DNA methylation data and the gene-expression data both contained 562 samples, comprised of 49 matched normal samples, 494 PTC samples, 11 metastatic thyroid carcinoma samples, and eight samples of other types of thyroid cancers. We eliminated batch effects between these 562 samples. In total, 49 PTC samples and 49 matched normal samples comprised the training set, with the remaining 445 PTC samples used as the testing set. Three samples were excluded because they did not contain survival information. The final study included 442 PTC patients in the testing set (Table 1). Eleven metastatic thyroid carcinoma samples and eight samples of other types of thyroid cancers were excluded.
Table 1.
Clinical Information on PTC Patients
| Characteristics | Training Set (n = 49) | Testing Set (n = 442) |
|---|---|---|
| State | ||
| Living | 45 | 431 |
| Dead | 4 | 11 |
| Survival (years) | ||
| Mean ± SD | 4.22 ± 3.52 | 2.46 ± 2.37 |
| Gender | ||
| Male | 14 | 117 |
| Female | 35 | 325 |
| Age (years) | ||
| Mean ± SD | 45.29 ± 17.22 | 47.57 ± 15.72 |
| Stage | ||
| I | 30 | 246 |
| II | 5 | 47 |
| III | 11 | 99 |
| IV | 3 | 50 |
| Histological Type | ||
| Thyroid papillary carcinoma | ||
| Classical/usual | 42 | 313 |
| Tall cell (≥50% tall cell features) | 3 | 33 |
| Follicular (≥99% follicular patterned) | 4 | 96 |
All of the CpG sites were from the Illumina Infinium HumanMethylation450 BeadChip assay. Raw data (level 3 data) contained all CpG sites in the gene sequence and all CpG sites in the promoter of the analyzed gene. For a specific CpG site, we calculated the correlations with expression of the nearest gene. In total, 203,015 differentially methylated CpG sites were identified from the training set, and 7,541 differentially methylated CpG sites were significantly related to gene expression (false discovery rate [FDR]-corrected p value < 0.05) and included 4,997 genes. There were 3,673 hypermethylated sites and 3,868 hypomethylated sites. More than 50% of the hypomethylated sites were negatively related to gene expression (Figure S1A), and more than 50% of the hypermethylated sites were negatively related to gene expression (Figure S1B). A total of 2,035 genes with the proportion of the hypomethylated sites associated with gene expression were greater than 9.60%, accounting for more than 50%, and the expression of seven genes was influenced by all of their hypomethylated sites (Figure S1C). A total of 2,082 genes with the proportion of the hypermethylated sites associated with gene expression were greater than 9.22%, accounting for more than 50%, and the expression of three genes was influenced by all of their hypermethylated sites (Figure S1D). The hypomethylated sites showed significant regulatory effects on gene expression (Figure S1E), and the hypermethylated sites also showed significant regulatory effects on gene expression (Figure S1F).
Identification of the Genes Related to CIN
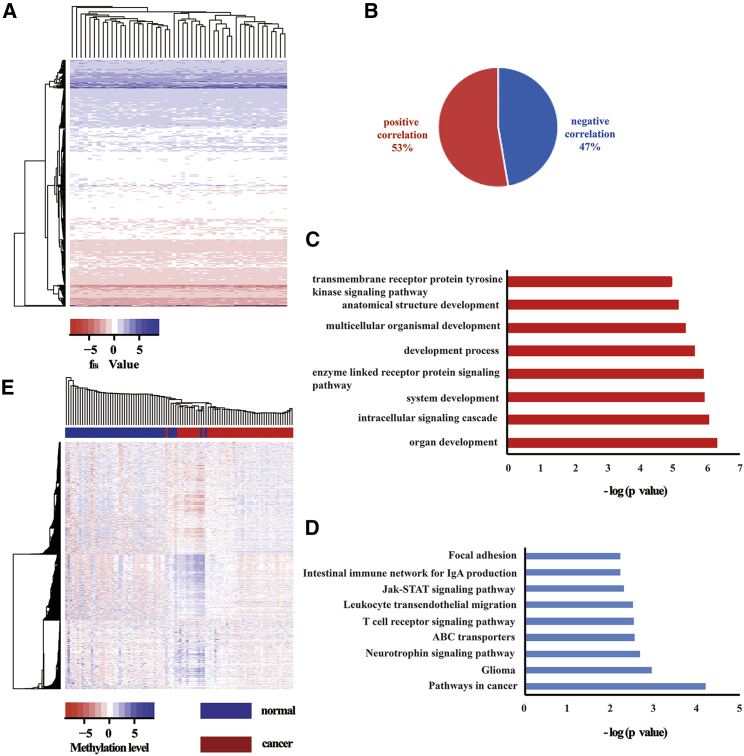
The methylation pattern score (score value) was used to describe the maximal regulatory effect of DNA methylation on gene expression. The distribution of the score value was consistent with the normal distribution, which was the same as the gene expression (Figure S2). We therefore characterized the CIN based on the score value using Student’s t statistics. The fBi was the net deviation in the score value contained in each chromosomal region relative to the remainder of the sampled score value. The results of clustering analyses showed that most of the bands had CIN (Figure 1A). As a measure of overall CIN, the MFA of a sample was defined as the sum of the magnitudes of its fBi features. In total, the DNA methylation patterns of 1,239 genes from preselected 4,997 genes were significantly related to the MFA (Table S1), and 53% of them showed a positive correlation, indicating the higher the gene score value, the higher the CIN (Figure 1B). A total of 572 of them were hypermethylation phenotypes (Table S2), and 667 of them were hypomethylation phenotypes (Table S3).
Figure 1.
Analyses of Papillary Thyroid Carcinoma Data
(A) Two-way hierarchical clustering of the fBi value in papillary thyroid carcinoma tissue samples. The blue area was >0, and the red area was <0. (B) In total, DNA methylation patterns of 1,239 genes were highly related to chromosomal instability (CIN), and 653 (53% of the total) were positively correlated genes. In total, 586 (47% of the total) were negatively correlated genes. (C) Gene ontology (GO) functional enrichment analyses for DNA methylation patterns of 1,239 genes were highly related to CIN using DAVID. (D) The Kyoto Encyclopedia of Genes and Genomes pathway enrichment analyses for DNA methylation patterns of 1,239 genes were highly related to CIN using DAVID. (E) Two-way hierarchical clustering of DNA methylation patterns of 1,239 genes were highly related to CIN in papillary thyroid carcinoma tissue samples and normal tissue samples. The blue area was >0, and the red area was <0.
Gene ontology (GO) functional enrichment analyses (Figure 1C) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses (Figure 1D) showed that the 1,239 genes were significantly related to organ development and were enriched in pathways that were related to cancer. The DNA methylation patterns of these genes were significantly related to the MFA and significantly differentiated between normal samples and cancer samples (Figure 1E).
Differentially Methylated Markers Associated with PTC Clinical Prognoses
We used the log-rank test to analyze the DNA methylation patterns of 1,239 genes that had high correlations with CIN, with 28 genes being left. The DNA methylation patterns of thirteen genes that had high correlations with CIN were significant (p < 0.05) using univariate Cox regression analyses. The hazard ratios (HRs) and 95% confidence intervals (CIs) of the clinical parameters for mortality were calculated using univariate Cox proportional hazard model analyses. The Kaplan-Meier method was used to estimate the overall survival times of patients. The DNA methylation patterns of four genes, TMEM18, UBAC2, ELOVL2, and ALMS1P, which had high correlations with CIN, were finally identified as prognosis markers of PTC. We developed four distinct risk scores, each of them based on the methylation pattern of one of four genes. The risk score formula for each patient was calculated as follows: risk score = (−20.05 × TMEM18), (−41.68 × UBAC2), (−6.20807 × ELOVL2), and (21.25 × ALMS1P). If the regression coefficient estimated by the univariate Cox proportional hazards model > 0, then the hyper-methylation of the risk gene was bad for the survival time. We subdivided the PTC patients into high-risk and low-risk groups by using the median of the risk scores. The HRs (95% CI) of TMEM18, UBAC2, ELOVL2, and ALMS1P were 1.951 × 10−9 (2.351e−17-0.1619), 7.909 × 10−19 (3.658e−33-0.000171), 0.002013 (5.646e−06-0.7177), and 1.7 × 109 (1.075-2.687e+18), respectively (Table 2).
Table 2.
Prognosis Markers of PTC Patients
| Variable | HR (95% CI) | Regression Coefficient | p Value |
|---|---|---|---|
| TMEM18 | 1.951 × 10-9(2.351e−17-0.1619) | −20.05 | 0.0311 |
| UBAC2 | 7.909 × 10-19(3.658e−33-0.000171) | −41.68 | 0.0133 |
| ELOVL2 | 0.002013(5.646e−06-0.7177) | −6.208070 | 0.0384 |
| ALMS1P | 1.7 × 109(1.075-2.687e+18) | 21.25 | 0.0492 |
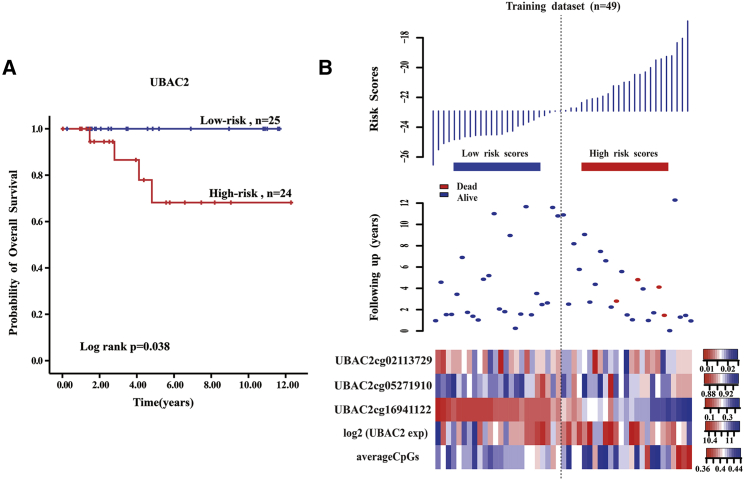
The DNA methylation pattern of UBAC2 that had a high correlation with CIN significantly predicted the survival of PTC patients in the training set (Figure 2A). The 5-year survival percentage of the high-risk score patients was 68.2% ± 13.6% and was less than that of the low-risk score patients (100%, p = 0.038). From the low-risk group to the high-risk group, the methylation levels of the UBAC2 cg16941122 site showed a significant upward trend and had a strong linear relationship with the risk index, although the expression levels of UBAC2 and the average methylation levels had no obvious trend (Figure 2B). Furthermore, a higher UBAC2 risk score was associated with a worse PTC patient prognosis.
Figure 2.
The UBAC2 Risk Score Model Predicts Overall Survival of Papillary Thyroid Carcinoma Patients in the Training Dataset
(A) Kaplan-Meier analyses for overall survival of patients with high-risk or low-risk scores. The p value was calculated using the two-sided log-rank test. (B) DNA methylation pattern, expression levels, and average methylation levels of the prognostic UBAC2 that correlated with patients’ survival status and increased risk scores.
The DNA methylation pattern of TMEM18 that had a high correlation with the CIN significantly predicted the survival of PTC patients in the training set. The 5-year survival percentage of the high-risk score patients was 59.8% ± 16.3% and was significantly lower than that of the low-risk score patients (100%, p = 0.01) (Figure S3). The DNA methylation pattern of ALMS1P that had a high correlation with CIN significantly predicted the survival of PTC patients in the training set. The 5-year survival percentage of the high-risk score patients was 67.6% ± 14.2% and was significantly lower than that of the low-risk score patients (100%, p = 0.041) (Figure S4). Therefore, a higher gene risk score predicted a worse PTC patient prognosis.
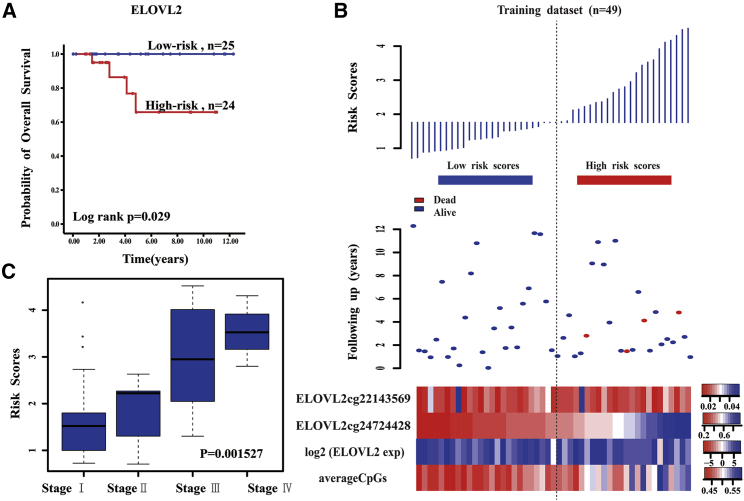
The DNA methylation pattern of ELOVL2 that had a high correlation with CIN significantly predicted the survival of PTC patients in the training set (Figure 3A). The 5-year survival percentage of the high-risk score patients was 65.8% ± 14.6% and was significantly lower than that of the low-risk score group (100%, p = 0.029). From the low-risk group to the high-risk group, the methylation levels of the ELOVL2 cg24724428 site showed a significant upward trend and had a strong linear relationship with the risk index, although the expression levels of ELOVL2 and the average methylation levels had no obvious trend (Figure 3B). Moreover, the ELOVL2 risk score was significantly different in the four stages of PTC (Kruskal-Wallis test, p = 0.001527), suggesting that the DNA methylation pattern of ELOVL2 that had a high correlation with CIN was related to the progress of PTC (Figure 3C). A higher ELOVL2 risk score correlated with a worse PTC patient prognosis, further suggesting that the ELOVL2 score value that had a high correlation with CIN significantly influenced the patient’s clinical condition, progression of the disease, and survival time.
Figure 3.
The ELOVL2 Risk Score Model Predicts Overall Survival of Papillary Thyroid Carcinoma Patients in the Training Dataset
(A) Kaplan-Meier analyses for overall survival of patients with high-risk or low-risk scores. The p value was calculated using the two-sided log-rank test. (B) DNA methylation pattern, expression levels, and average methylation levels of the prognostic ELOVL2 that correlated with patients’ survival status and increased risk scores. (C) Box plot of the ELOVL2 risk scores in the four stages of papillary thyroid carcinoma.
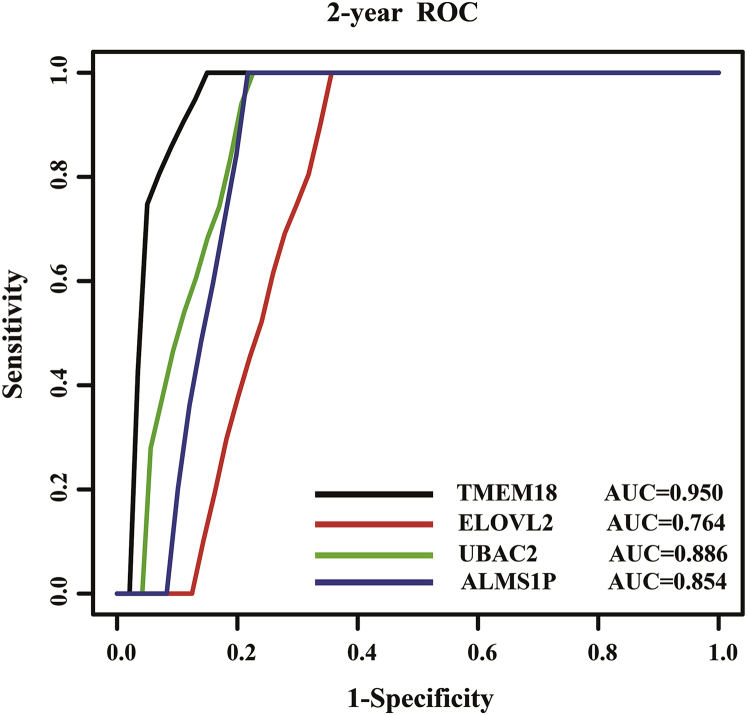
We performed a time-dependent receiver-operating characteristic (ROC) curve analysis to compare the sensitivity and specificity for survival predictions among the DNA methylation patterns of these four genes. The area under the ROC curve (AUC) value was obtained from the ROC analyses and was compared among the DNA methylation patterns of these four genes. The AUC values of TMEM18, UBAC2, ELOVL2, and ALMS1P were 0.95, 0.886, 0.764, and 0.854, respectively (Figure 4).
Figure 4.
Receiver-Operated Characteristic (ROC) Analyses of the Sensitivity and Specificity for Survival Prediction among the DNA Methylation Patterns of the Four Genes
The time-dependent ROC curve was used to evaluate the prognostic performance for survival predictions. The performance comparison was assessed among the four genes by calculating the area under the ROC curves (AUC) in the training dataset.
Verification of the Testing Set
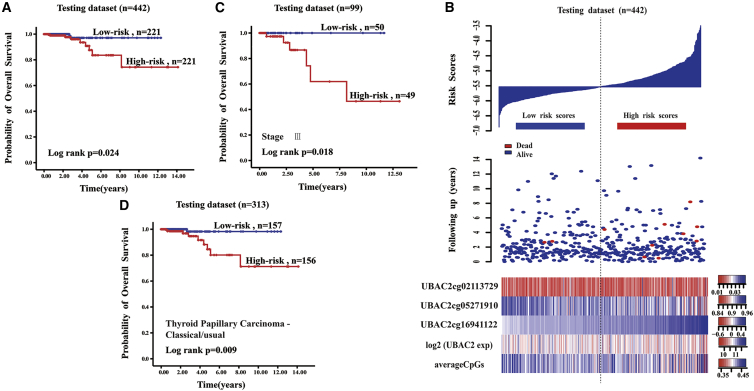
The testing set was used to evaluate the reproducibility and availability of these four genes in a prognostic model. The DNA methylation patterns of ELOVL2 and UBAC2 were obtained using independent cancer samples that not only associated with CIN but could also be used to predict the prognosis of PTC. Moreover, UBAC2 and ELOVL2 were hypomethylation phenotypes. The DNA methylation pattern of UBAC2 that had a high correlation with CIN predicted the survival of PTC patients in the testing set (Figure 5A). The 5-year survival percentage of the high-risk score patients was 83.6% ± 6.2%, which was significantly lower than that of the low-risk score group (97.1% ± 2%, p = 0.024). From the low-risk group to the high-risk group, the methylation levels of the UBAC2 cg16941122 site showed a significant upward trend and had a strong linear relationship with the risk index, although the expression levels of UBAC2 and the average methylation levels had no obvious trend (Figure 5B). The samples from the testing set were subgrouped based on the tumor stage, and the survival times of patients from the high-risk score group were significantly different from that of the low-risk score group in stage III of PTC (p = 0.018) (Figure 5C). The survival times of patients from the high-risk score group were the same as those from the low-risk score group in stages I, II, and IV of PTC (p = 0.39, 0.355, 0.137, respectively). We subgrouped the testing samples based on histological type, and the survival times of the patients in the high-risk score group were significantly different from those of the low-risk score group in the thyroid papillary carcinoma classical/usual group (p = 0.009) (Figure 5D). The thyroid papillary carcinoma tall cell (≥50% tall cell features) histological group contained 33 samples, and there were no samples from patients who had died. Therefore, we could not perform survival analyses. The survival times of the patients in the high-risk score group were the same as those from the low-risk score group in the thyroid papillary carcinoma-follicular (≥99% follicular patterned) histological group (p = 0.366). A higher UBAC2 risk score predicted a worse PTC patient prognosis.
Figure 5.
The UBAC2 Risk Score Model Predicts Overall Survival of Papillary Thyroid Carcinoma Patients in the Testing Dataset
(A) Kaplan-Meier analyses for overall survival of patients with high-risk or low-risk scores. The p value was calculated using the two-sided log-rank test. (B) DNA methylation pattern, expression level, and average methylation level of the prognostic UBAC2 that correlated with patients’ survival status and increased risk scores. (C) Kaplan-Meier analyses for overall survival of patients in the third stage of papillary thyroid carcinoma. The p value was calculated using the two-sided log-rank test. (D) Kaplan-Meier analyses for overall survival of patients with thyroid papillary carcinoma-classical/usual histological types. The p value was calculated using the two-sided log-rank test.
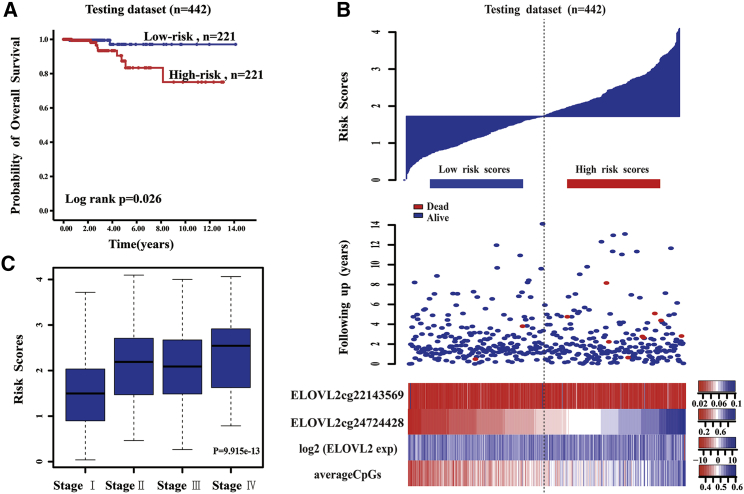
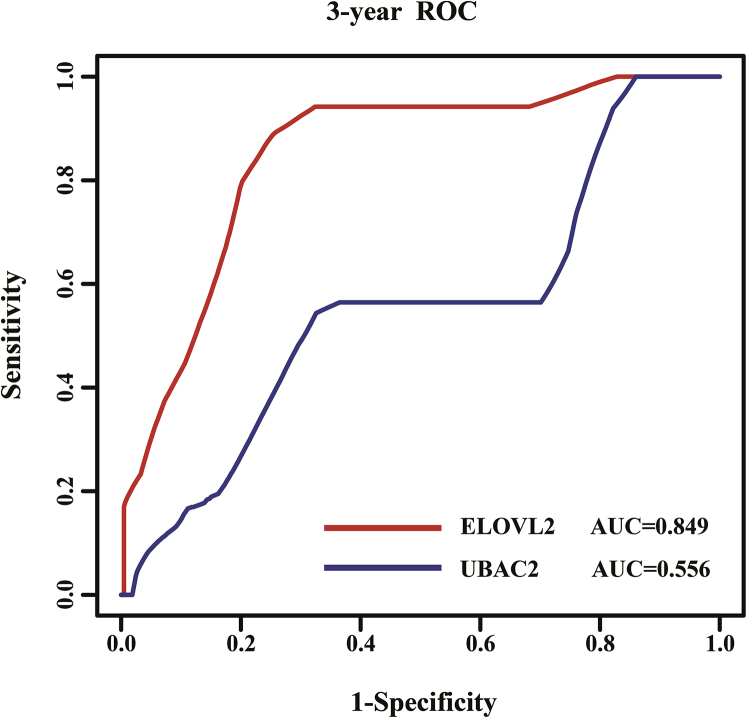
The DNA methylation pattern of ELOVL2 that had a high correlation with the CIN significantly predicted the survival of PTC patients in the testing set (Figure 6A). The 5-year survival percentage of the high-risk score patients was 83.4% ± 6.1% and was significantly lower than that of the low-risk score group (97.1% ± 2.4%; p = 0.026). From the low-risk group to the high-risk group, the methylation levels of the ELOVL2 cg24724428 site showed a significant upward trend and had a strong linear relationship with the risk index, although the expression levels of ELOVL2 and the average methylation levels had no obvious trend (Figure 6B). The samples from the testing set were subgrouped based on the tumor stage, and the survival times of patients from the high-risk score group were the same as those of the low-risk score group in stages I, II, III, and IV of PTC (p = 0.324, 0.206, 0.19, and 0.05, respectively). We subgrouped the testing samples based on histological type, and the survival times of the patients in the high-risk score group were the same as those from the low-risk score group in the thyroid papillary carcinoma classical/usual group (p = 0.078). The survival times of the patients in the high-risk score group were the same as those of the low-risk score group in the thyroid papillary carcinoma-follicular (≥99% follicular pattern) histological group (p = 0.317). Moreover, the ELOVL2 risk score was significantly different in the four stages of PTC (Kruskal-Wallis test; p = 9.915 × 10−13) (Figure 6C), suggesting that the DNA methylation pattern of ELOVL2 that had a high correlation with CIN was related to the progress of PTC. A higher ELOVL2 risk score was correlated with a worse PTC patient prognosis, indicating that the ELOVL2 score value that had a high correlation with CIN significantly influenced the patient’s clinical condition, progression of disease, and survival time. The AUC values of ELOVL2 and UBAC2 were 0.849 and 0.556, respectively (Figure 7).
Figure 6.
The ELOVL2 Risk Score Model Predicts Overall Survival of Papillary Thyroid Carcinoma Patients in the Testing Dataset
(A) Kaplan-Meier analyses for overall survival of patients with high-risk or low-risk scores. The P value was calculated using the two-sided log-rank test. (B) DNA methylation pattern, expression level, and average methylation level of the prognostic ELOVL2 that correlated with the patients’ survival status and increased risk scores. (C) Box plot of the ELOVL2 risk score in the four stages of papillary thyroid carcinoma.
Figure 7.
Receiver-Operated Characteristic (ROC) Analyses of the Sensitivity and Specificity for Survival Prediction between the DNA Methylation Patterns of the Two Genes
The time-dependent ROC curve was used to evaluate the prognostic performance for survival predictions. The performance comparison was assessed between the two genes by calculating the area under the ROC curves (AUC) in the testing dataset.
Discussion
To predict PTC patient clinical prognoses, tumor node metastasis (TNM) staging; patient age, histologic grade of the tumor, tumor extent (extrathyroidal invasion or distant metastases), and size of the primary tumor (AGES) scoring; patient age, presence of distant metastases, extent and size of the primary tumor (AMES) scoring; and metastasis, patient age, completeness of resection, local invasion, and tumor size (MACIS) scoring have been used.32, 33, 34, 35 However, patients with similar clinical phenotypes do not have identical prognoses, suggesting that the present PTC prognostic evaluation system does not provide an accurate clinical prognosis for every patient.36, 37, 38, 39 The PTC prognostic evaluation system therefore needs improvement. The accuracy of PTC prognoses could be significantly improved by the use of molecular markers. Cancer patients with a higher CIN have a worse clinical prognosis, so CIN could be used to evaluate the clinical prognoses of tumor patients.40, 41, 42, 43 To improve the existing PTC prognostic evaluation system, it is important for PTC patient treatment to identify reliable CIN-related prognostic markers. Although CIN-related prognostic markers have been previously reported,9 the results differed. Therefore, more valid CIN-related prognostic markers are needed to improve the accuracy and credibility of the prognoses.
DNA methylation plays an important role in the regulation of gene expression and CIN. Based on PTC samples from TCGA database, we used the R Significance Analysis of Microarrays (SAM) package44 to identify 203,015 differentially methylated CpG sites between PTC and normal samples. Then, we used multiple regression analyses to obtain 7,541 methylation patterns of CpG sites with the strongest correlation with gene expressions. A total of 4,997 genes were obtained by combining the CpG sites, which were represented as featured DNA methylation patterns. The results showed that the distribution of DNA methylation patterns was consistent with the normal distribution, which was the same as gene expression. We subsequently developed a method to characterize CIN based on DNA methylation patterns of genes using the Student’s t statistics. PCC was used to evaluate the correlation between the DNA methylation patterns and the CIN of each gene. We found that 1,239 genes were highly associated with CIN. With the use of the log-rank test, univariate Cox regression analyses, and the Kaplan-Meier method, DNA methylation patterns of four genes, including TMEM18, UBAC2, ELOVL2, and ALMS1P, which had high correlations with CIN, provided good prognostic values for PTC. An independent test set was used to test the validity of the methylation risk score of the four genes. Finally, ELOVL2 and UBAC2 remained. In addition, the DNA methylation pattern of ELOVL2 was involved in different stages of PTC, indicating that the DNA methylation pattern of ELOVL2 with a high correlation with CIN significantly influenced the patient’s clinical condition, progression of disease, and survival time. The DNA methylation pattern associated with CIN may therefore be a good predictor of PTC survival.
Previous studies of ELOVL2 and UBAC2 emphasized their relationships with lipid metabolism and obesity.45, 46, 47, 48 González-Bengtsson et al.45 suggested that ELOVL2 played an important role in docosahexaenoic acid (DHA) synthesis. Kobayashi et al.46 reported that cells overexpressing ELOVL2 showed enhanced triacylglycerol synthesis and subsequent accumulation of lipid droplets. Pauter et al.47 suggested that hepatic DHA synthesis of ELOVL2, in addition to controlling de novo lipogenesis, also regulated lipid storage and fat mass expansion in an SREBP-1c-independent fashion. Tikhonenko et al.48 reported that a decrease in long-chain polyunsaturated fatty acids was associated with a decrease in the fatty acid elongases, ELOVL2 and ELOVL4, in diabetes, and additional studies showed that obesity increased the risk of thyroid cancer.49, 50, 51, 52 Han et al.49 reported that the morbidity of thyroid cancer in female patients was related to a high BMI. Hwang et al.50 suggested that weight gain and annual increases in obesity indicators in middle-aged adults increased the risk of developing PTC. Kim et al.51 reported that a higher BMI was associated with more aggressive tumor features, such as lymph node metastasis, lymphatic invasion, and tumor multiplicity in PTC patients. Oberman et al.52 reported that obesity was significantly associated with thyroid cancer, with BMI, in particular, a strong predictor of thyroid cancer. ELOVL2 was also significantly related to biosynthesis of unsaturated fatty acids, and UBAC2 was significantly related to protein localization to the endoplasmic reticulum. Therefore, DNA methylation patterns of ELOVL2 and UBAC2 were not only associated with CIN, but might also participate in the initiation and development of PTC.
Because of differences among individual patients, presently used prognostic indicators cannot accurately predict the prognosis of each patient. It is therefore difficult to evaluate the clinical prognoses of patients with similar clinical features. Our results showed that the DNA methylation patterns of ELOVL2 and UBAC2, which had high correlations with CIN, could be used to predict the prognosis of PTC. Aberrant DNA methylation was related to the risk of PTC, and the epigenetic markers associated with CIN may be used as predictors of PTC survival. The inclusion of these prognosis markers into the present PTC prognostic evaluation system could therefore assist the clinician in determining the prognoses of patients with similar clinical features and could provide a more appropriate therapeutic schedule for high-risk patients to enhance the efficacies of PTC treatments.
Conclusions
UBAC2 and ELOVL2, which had high correlations with CIN, provided good prognostic values for PTC. The DNA methylation pattern associated with CIN may therefore be a good predictor of PTC survival.
Materials and Methods
Acquisition of Gene Expression and DNA Methylation Data
With the use of the Illumina Infinium HumanMethylation450 BeadChip assay, raw data (level 3 data), raw UNC RNAseqV2 level 3 expression data, and clinical prognostic information for PTC were collected from TCGA (https://www.cancer.gov/tcga/).
Eliminating Batch Effects
In order to ensure the accuracy of experiments, we used the R Surrogate Variable Analysis (Bioconductor) package to eliminate batch effects of all of the DNA methylation data and the gene-expression data from all samples.
Identifying Differentially Methylated CpGs
To compare the differences of DNA methylation between cancer and normal samples in the training set, we used the R SAM44 package to identify differentially methylated CpG sites. To control for a FDR of the results, we used the Benjamini-Hochberg method to correct the p value obtained from the statistical test. The threshold for defining the differentially methylated CpG site involved a value of p < 0.05, and the differential level (delta beta value) between the cancer and normal samples was >0.1.
Identifying Differentially Methylated CpGs Associated with Gene Expression
For the differentially methylated CpG sites of the PTC samples from the training set, we used multiple regression coefficients to evaluate the correlation between DNA methylation and gene expression. The dependent variable was gene expression of a single gene. The independent variables were all CpG sites mapped to the gene, containing all CpG sites under the gene sequence and all CpG sites in the promoter of the analyzed gene. For a particular CpG site, we calculated correlations with the expression of the nearest gene. A value of p < 0.05 was identified as a significant methylation level highly related to gene expression.
Quantitation of the Regulatory Effect of DNA Methylation (Score Value)
With the consideration of the multiple CpGs mapped to the gene and the variability of the DNA methylation levels of multiple CpGs located in the same gene, the average methylation level may not reflect the real ability of DNA methylation to regulate gene expression. Multiple regression analyses were therefore used to quantify the regulatory competence of differential CpG sites and then to quantify the maximal regulatory effect of DNA methylation on gene expression. The methylation pattern score (score value) was defined31 as follows:
| (Equation 1) |
The j CpGs sites represented the significant methylation sites in multiple regression analyses (p < 0.05), αˆj (j = 1,2,…,j) represented the multiple regression coefficient of the jth CpG sites of the gene, and cgkj represented the methylation level of the jth CpG sites of the gene in the kth sample.
Calculating the CIN Score (MFA)
This study downloaded the cytoband coordinate file (GRCh37/hg19) from the UCSC Genome Bioinformatics and then mapped the genes to chromosomal sub-bands. If fewer than five genes were present in a given cytoband, we considered the statistical measure unreliable, and that cytoband was eliminated from further analysis. The fBi described the net deviation in score value contained in each chromosomal region relative to the remainder of the sampled score value:9
| (Equation 2) |
The μBi represented the average score value of all of the genes under sample i in the band, represented the variance of the score value of all the genes under sample i in the band, μGi represented the average score value of the rest of the genes under sample i, represented the variance of the score value of the rest of the genes under sample i, NB represented the number of genes in the band, and NG represented the number of the remaining genes.
As a measure of overall CIN, we defined the MFA of a sample as the sum of the magnitudes of its fBi features:9
| (Equation 3) |
Identification of Genes Related to MFA
PCC was used to evaluate the correlation between the score value and the MFA of each gene. In order to control for a FDR of the PCC, we adopted a permutation test to correct the p value of the statistical test. For each gene, the MFA was permutated 1,000 times to calculate its PCC value, and if the value of p was <0.05, this gene was identified as highly related to MFA as follows:
| (Equation 4) |
where c was the number of PCC square values that were no less than the actual PCC square value of the gene from the 1,000 permutations.
Enrichment Analyses for the GO and KEGG Pathways
To analyze further the biological significance of the genes related to CIN, we used Database for Annotation, Visualization and Integrated Discovery (DAVID) software to perform GO function analyses for the genes related to CIN.53 The Fisher exact test with multiple test corrections (FDR < 0.05) was used to obtain significant GO terms associated with PTC. We acquired the KEGG pathway terms using the same method.
Prognosis Analyses
The log-rank test was used to obtain p values and to identify a subset of genes for which a score value that had high correlations with CIN showed significant differences between the high and low groups. The high and low groups were groups with high and low score values, grouped by their median values. The survival times were compared between these two groups. Genes with p < 0.05 were used in the study. The p values were uncorrected p values. Univariate Cox regression analyses were performed to assess the survival prognosis capabilities of the selected gene set using the overall survival time as a dependent variable. The HRs and 95% CIs of the clinical parameters for mortality were calculated using the univariate Cox proportional hazard model. The risk score formula for each patient was calculated as follows:
| (Equation 5) |
where k was the kth sample, i denoted the feature genes filtered by the univariate Cox proportional hazards models, and ci was the regression coefficient estimated by the univariate Cox proportional hazards model. The 5-year overall survival for each score value scoring group (high versus low) was calculated using the Kaplan-Meier method, and the statistical significance was assessed using the log-rank test. The significance level of all statistical tests was p < 0.05. We performed time-dependent ROC curve analyses to compare the sensitivities and specificities for survival predictions between the predicted genes. The ROC AUC values were obtained from ROC analyses and were compared between the selected genes.
In order to verify the reproducibility and accuracy of the gene prognostic model, as predicted in the training set, we used the testing set. The regression coefficients and the thresholds of risk scores derived from the training set were directly applied to the testing set, and then the patients in the testing set were divided into high-risk and low-risk groups. The evaluation of survival times and the comparison of differences between the two groups were the same as that of the training set.
Author Contributions
H.Q. conceived and designed the experiments. J.H., M.C., and Q.F. acquired the experiment data. J.H., M.C., and Y.Z. performed the study. J.H., M.C., and Y.W. carried out the data analysis. J.H. and J.E. wrote the manuscript. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was funded by the National Natural Science Foundation of China (grant number 81673108), the Science and Technology Innovation Talent Research Foundation of Harbin (grant number 2016RAXYJ088), and the Nurture Research Fund from The Fourth Affiliated Hospital of Harbin Medical University (grant number HYDSYPY201910).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.09.027.
Supplemental Information
References
- 1.Bădulescu I.C., Bărbuş E., Piciu D. Circulating tumor cells in thyroid carcinoma - the prognostic role of this biomarker. Review of the literature. Clujul Med. 2017;90:256–261. doi: 10.15386/cjmed-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang H., Teng X., Liu Z., Zhang L., Liu Z. Gene expression profile analyze the molecular mechanism of CXCR7 regulating papillary thyroid carcinoma growth and metastasis. J. Exp. Clin. Cancer Res. 2015;34:16. doi: 10.1186/s13046-015-0132-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang P., Pei R., Lu Z., Rao X., Liu B. Methylation of p16 CpG islands correlated with metastasis and aggressiveness in papillary thyroid carcinoma. J. Chin. Med. Assoc. 2013;76:135–139. doi: 10.1016/j.jcma.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 4.Lewiński A., Adamczewski Z. Papillary thyroid carcinoma: a cancer with an extremely diverse genetic background and prognosis. Pol. Arch. Intern. Med. 2017;127:388–389. doi: 10.20452/pamw.4058. [DOI] [PubMed] [Google Scholar]
- 5.Hay I.D., Thompson G.B., Grant C.S., Bergstralh E.J., Dvorak C.E., Gorman C.A., Maurer M.S., McIver B., Mullan B.P., Oberg A.L. Papillary thyroid carcinoma managed at the Mayo Clinic during six decades (1940-1999): temporal trends in initial therapy and long-term outcome in 2444 consecutively treated patients. World J. Surg. 2002;26:879–885. doi: 10.1007/s00268-002-6612-1. [DOI] [PubMed] [Google Scholar]
- 6.Cancer Genome Atlas Research Network Integrated genomic characterization of papillary thyroid carcinoma. Cell. 2014;159:676–690. doi: 10.1016/j.cell.2014.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma M., Yu N. Over-Expression of TBL1XR1 Indicates Poor Prognosis of Serous Epithelial Ovarian Cancer. Tohoku J. Exp. Med. 2017;241:239–247. doi: 10.1620/tjem.241.239. [DOI] [PubMed] [Google Scholar]
- 8.Wang M., Zhang G., Wang Y., Ma R., Zhang L., Lv H., Fang F., Kang X. DHX32 expression is an indicator of poor breast cancer prognosis. Oncol. Lett. 2017;13:942–948. doi: 10.3892/ol.2016.5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carter S.L., Eklund A.C., Kohane I.S., Harris L.N., Szallasi Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat. Genet. 2006;38:1043–1048. doi: 10.1038/ng1861. [DOI] [PubMed] [Google Scholar]
- 10.Bastians H. Causes of Chromosomal Instability. Recent Results Cancer Res. 2015;200:95–113. doi: 10.1007/978-3-319-20291-4_5. [DOI] [PubMed] [Google Scholar]
- 11.Carbone L., Chavez S.L. Mammalian pre-implantation chromosomal instability: species comparison, evolutionary considerations, and pathological correlations. Syst Biol Reprod Med. 2015;61:321–335. doi: 10.3109/19396368.2015.1073406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meena J., Rudolph K.L., Günes C. Telomere Dysfunction, Chromosomal Instability and Cancer. Recent Results Cancer Res. 2015;200:61–79. doi: 10.1007/978-3-319-20291-4_3. [DOI] [PubMed] [Google Scholar]
- 13.Wu Z.H. Phenotypes and genotypes of the chromosomal instability syndromes. Transl. Pediatr. 2016;5:79–83. doi: 10.21037/tp.2016.03.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawano H., Saeki H., Kitao H., Tsuda Y., Otsu H., Ando K., Ito S., Egashira A., Oki E., Morita M. Chromosomal instability associated with global DNA hypomethylation is associated with the initiation and progression of esophageal squamous cell carcinoma. Ann. Surg. Oncol. 2014;21(Suppl 4):S696–S702. doi: 10.1245/s10434-014-3818-z. [DOI] [PubMed] [Google Scholar]
- 15.Nishida N., Kudo M., Nishimura T., Arizumi T., Takita M., Kitai S., Yada N., Hagiwara S., Inoue T., Minami Y. Unique association between global DNA hypomethylation and chromosomal alterations in human hepatocellular carcinoma. PLoS ONE. 2013;8:e72312. doi: 10.1371/journal.pone.0072312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rodriguez J., Frigola J., Vendrell E., Risques R.A., Fraga M.F., Morales C., Moreno V., Esteller M., Capellà G., Ribas M., Peinado M.A. Chromosomal instability correlates with genome-wide DNA demethylation in human primary colorectal cancers. Cancer Res. 2006;66:8462–9468. doi: 10.1158/0008-5472.CAN-06-0293. [DOI] [PubMed] [Google Scholar]
- 17.Suzuki K., Suzuki I., Leodolter A., Alonso S., Horiuchi S., Yamashita K., Perucho M. Global DNA demethylation in gastrointestinal cancer is age dependent and precedes genomic damage. Cancer Cell. 2006;9:199–207. doi: 10.1016/j.ccr.2006.02.016. [DOI] [PubMed] [Google Scholar]
- 18.Bird A. Perceptions of epigenetics. Nature. 2007;447:396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- 19.Jaenisch R., Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 20.Laird P.W. Principles and challenges of genomewide DNA methylation analysis. Nat. Rev. Genet. 2010;11:191–203. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- 21.Taby R., Issa J.P. Cancer epigenetics. CA Cancer J. Clin. 2010;60:376–392. doi: 10.3322/caac.20085. [DOI] [PubMed] [Google Scholar]
- 22.Lu X.X., Ge M.H., Ling Z.Q., Hu S.S., Xu J., Zheng C.M., Tan Z., Chen C. [Aberrant methylation of hMLH1 gene promoter in papillary thyroid cancer and its clinical significance] Zhonghua Zhong Liu Za Zhi. 2013;35:833–836. [PubMed] [Google Scholar]
- 23.Shou F., Xu F., Li G., Zhao Z., Mao Y., Yang F., Wang H., Guo H. RASSF1A promoter methylation is associated with increased risk of thyroid cancer: a meta-analysis. OncoTargets Ther. 2017;10:247–257. doi: 10.2147/OTT.S124417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang D., Cui W., Wu X., Qu Y., Wang N., Shi B., Hou P. RUNX3 site-specific hypermethylation predicts papillary thyroid cancer recurrence. Am. J. Cancer Res. 2014;4:725–737. [PMC free article] [PubMed] [Google Scholar]
- 25.El-Tahan R.R., Ghoneim A.M., El-Mashad N. TNF-α gene polymorphisms and expression. Springerplus. 2016;5:1508. doi: 10.1186/s40064-016-3197-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fujiuchi N., Matoba N., Matsuda R. Environment Control to Improve Recombinant Protein Yields in Plants Based on Agrobacterium-Mediated Transient Gene Expression. Front. Bioeng. Biotechnol. 2016;4:23. doi: 10.3389/fbioe.2016.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rivas H.G., Schmaling S.K., Gaglia M.M. Shutoff of Host Gene Expression in Influenza A Virus and Herpesviruses: Similar Mechanisms and Common Themes. Viruses. 2016;8:102. doi: 10.3390/v8040102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salminen A., Kauppinen A., Kaarniranta K. AMPK/Snf1 signaling regulates histone acetylation: Impact on gene expression and epigenetic functions. Cell. Signal. 2016;28:887–895. doi: 10.1016/j.cellsig.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 29.Judson H., Stewart A., Leslie A., Pratt N.R., Baty D.U., Steele R.J., Carey F.A. Relationship between point gene mutation, chromosomal abnormality, and tumour suppressor gene methylation status in colorectal adenomas. J. Pathol. 2006;210:344–350. doi: 10.1002/path.2044. [DOI] [PubMed] [Google Scholar]
- 30.Schultz M.D., He Y., Whitaker J.W., Hariharan M., Mukamel E.A., Leung D., Rajagopal N., Nery J.R., Urich M.A., Chen H. Human body epigenome maps reveal noncanonical DNA methylation variation. Nature. 2015;523:212–216. doi: 10.1038/nature14465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang M., Zhang S., Wen Y., Wang Y., Wei Y., Liu H., Zhang D., Su J., Wang F., Zhang Y. DNA Methylation Patterns Can Estimate Nonequivalent Outcomes of Breast Cancer with the Same Receptor Subtypes. PLoS ONE. 2015;10:e0142279. doi: 10.1371/journal.pone.0142279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chruścik A., Lam A.K. Clinical pathological impacts of microRNAs in papillary thyroid carcinoma: A crucial review. Exp. Mol. Pathol. 2015;99:393–398. doi: 10.1016/j.yexmp.2015.08.013. [DOI] [PubMed] [Google Scholar]
- 33.Gong W., Yang S., Yang X., Guo F. Blood preoperative neutrophil-to-lymphocyte ratio is correlated with TNM stage in patients with papillary thyroid cancer. Clinics (São Paulo) 2016;71:311–314. doi: 10.6061/clinics/2016(06)04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee Y.S., Kim Y., Jeon S., Bae J.S., Jung S.L., Jung C.K. Cytologic, clinicopathologic, and molecular features of papillary thyroid carcinoma with prominent hobnail features: 10 case reports and systematic literature review. Int. J. Clin. Exp. Pathol. 2015;8:7988–7997. [PMC free article] [PubMed] [Google Scholar]
- 35.Pillai S., Gopalan V., Smith R.A., Lam A.K. Diffuse sclerosing variant of papillary thyroid carcinoma--an update of its clinicopathological features and molecular biology. Crit. Rev. Oncol. Hematol. 2015;94:64–73. doi: 10.1016/j.critrevonc.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 36.Ito Y., Miyauchi A., Kihara M., Kobayashi K., Miya A. Prognostic values of clinical lymph node metastasis and macroscopic extrathyroid extension in papillary thyroid carcinoma. Endocr. J. 2014;61:745–750. doi: 10.1507/endocrj.ej14-0138. [DOI] [PubMed] [Google Scholar]
- 37.Ito Y., Miyauchi A., Kobayashi K., Kihara M., Miya A. Static and dynamic prognostic factors of papillary thyroid carcinoma. Endocr. J. 2014;61:1145–1151. doi: 10.1507/endocrj.EJ14-0303. [DOI] [PubMed] [Google Scholar]
- 38.Ito Y., Miyauchi A., Kobayashi K., Miya A. Prognosis and growth activity depend on patient age in clinical and subclinical papillary thyroid carcinoma. Endocr. J. 2014;61:205–213. doi: 10.1507/endocrj.ej13-0403. [DOI] [PubMed] [Google Scholar]
- 39.Pelizzo M.R., Merante Boschin I., Toniato A., Pagetta C., Casal Ide E., Mian C., Rubello D. Diagnosis, treatment, prognostic factors and long-term outcome in papillary thyroid carcinoma. Minerva Endocrinol. 2008;33:359–379. [PubMed] [Google Scholar]
- 40.Genga K.R., Filho F.D., Ferreira F.V., de Sousa J.C., Studart F.S., Magalhães S.M., Heredia F.F., Pinheiro R.F. Proteins of the mitotic checkpoint and spindle are related to chromosomal instability and unfavourable prognosis in patients with myelodysplastic syndrome. J. Clin. Pathol. 2015;68:381–387. doi: 10.1136/jclinpath-2014-202728. [DOI] [PubMed] [Google Scholar]
- 41.Giaretti W., Monteghirfo S., Pentenero M., Gandolfo S., Malacarne D., Castagnola P. Chromosomal instability, DNA index, dysplasia, and subsite in oral premalignancy as intermediate endpoints of risk of cancer. Cancer Epidemiol. Biomarkers Prev. 2013;22:1133–1141. doi: 10.1158/1055-9965.EPI-13-0147. [DOI] [PubMed] [Google Scholar]
- 42.Krem M.M., Press O.W., Horwitz M.S., Tidwell T. Mechanisms and clinical applications of chromosomal instability in lymphoid malignancy. Br. J. Haematol. 2015;171:13–28. doi: 10.1111/bjh.13507. [DOI] [PubMed] [Google Scholar]
- 43.Taiakina D., Dal Pra A., Bristow R.G. Intratumoral hypoxia as the genesis of genetic instability and clinical prognosis in prostate cancer. Adv. Exp. Med. Biol. 2014;772:189–204. doi: 10.1007/978-1-4614-5915-6_9. [DOI] [PubMed] [Google Scholar]
- 44.Li J., Tibshirani R. Finding consistent patterns: a nonparametric approach for identifying differential expression in RNA-Seq data. Stat. Methods Med. Res. 2013;22:519–536. doi: 10.1177/0962280211428386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.González-Bengtsson A., Asadi A., Gao H., Dahlman-Wright K., Jacobsson A. Estrogen Enhances the Expression of the Polyunsaturated Fatty Acid Elongase Elovl2 via ERα in Breast Cancer Cells. PLoS ONE. 2016;11:e0164241. doi: 10.1371/journal.pone.0164241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kobayashi T., Zadravec D., Jacobsson A. ELOVL2 overexpression enhances triacylglycerol synthesis in 3T3-L1 and F442A cells. FEBS Lett. 2007;581:3157–3163. doi: 10.1016/j.febslet.2007.05.081. [DOI] [PubMed] [Google Scholar]
- 47.Pauter A.M., Olsson P., Asadi A., Herslöf B., Csikasz R.I., Zadravec D., Jacobsson A. Elovl2 ablation demonstrates that systemic DHA is endogenously produced and is essential for lipid homeostasis in mice. J. Lipid Res. 2014;55:718–728. doi: 10.1194/jlr.M046151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tikhonenko M., Lydic T.A., Wang Y., Chen W., Opreanu M., Sochacki A., McSorley K.M., Renis R.L., Kern T., Jump D.B. Remodeling of retinal Fatty acids in an animal model of diabetes: a decrease in long-chain polyunsaturated fatty acids is associated with a decrease in fatty acid elongases Elovl2 and Elovl4. Diabetes. 2010;59:219–227. doi: 10.2337/db09-0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Han J.M., Kim T.Y., Jeon M.J., Yim J.H., Kim W.G., Song D.E., Hong S.J., Bae S.J., Kim H.K., Shin M.H. Obesity is a risk factor for thyroid cancer in a large, ultrasonographically screened population. Eur. J. Endocrinol. 2013;168:879–886. doi: 10.1530/EJE-13-0065. [DOI] [PubMed] [Google Scholar]
- 50.Hwang Y., Lee K.E., Park Y.J., Kim S.J., Kwon H., Park D.J., Cho B., Choi H.C., Kang D., Park S.K. Annual Average Changes in Adult Obesity as a Risk Factor for Papillary Thyroid Cancer: A Large-Scale Case-Control Study. Medicine (Baltimore) 2016;95:e2893. doi: 10.1097/MD.0000000000002893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim S.H., Park H.S., Kim K.H., Yoo H., Chae B.J., Bae J.S., Jung S.S., Song B.J. Correlation between obesity and clinicopathological factors in patients with papillary thyroid cancer. Surg. Today. 2015;45:723–729. doi: 10.1007/s00595-014-0984-3. [DOI] [PubMed] [Google Scholar]
- 52.Oberman B., Khaku A., Camacho F., Goldenberg D. Relationship between obesity, diabetes and the risk of thyroid cancer. Am. J. Otolaryngol. 2015;36:535–541. doi: 10.1016/j.amjoto.2015.02.015. [DOI] [PubMed] [Google Scholar]
- 53.Huang W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.