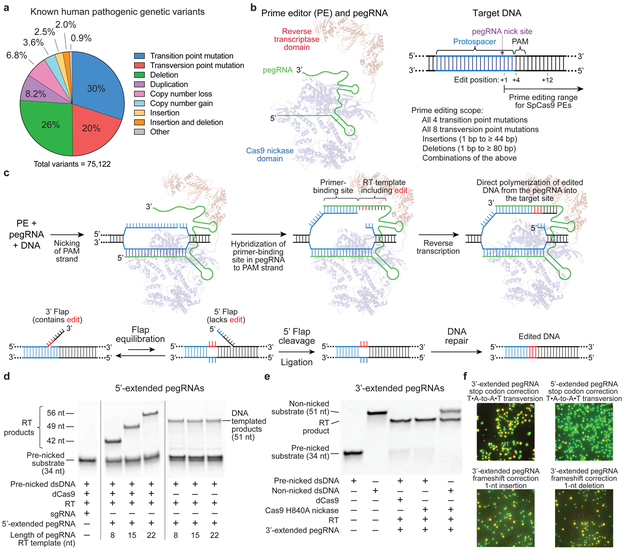
Figure 1. Overview of prime editing and feasibility studies in vitro and in yeast cells.
(a) The 75,122 known pathogenic human genetic variants in ClinVar (accessed July, 2019), classified by type. (b) A prime editing complex consists of a prime editor (PE) protein containing an RNA-guided DNA-nicking domain, such as Cas9 nickase, fused to a reverse transcriptase domain and complexed with a prime editing guide RNA (pegRNA). The PE:pegRNA complex enables a variety of precise DNA edits at a wide range of positions. (c) The PE:pegRNA complex binds the target DNA and nicks the PAM-containing strand. The resulting 3’ end hybridizes to the primer-binding site, then primes reverse transcription of new DNA containing the desired edit using the RT template of the pegRNA. Equilibration between the edited 3’ flap and the unedited 5’ flap, cellular 5’ flap cleavage and ligation, and DNA repair results in stably edited DNA. (d) In vitro primer extension assays with 5’-extended pegRNAs, pre-nicked dsDNA substrates containing 5’-Cy5 labeled PAM strands, dCas9, and a commercial M-MLV RT variant (RT, Superscript III). dCas9 was complexed with pegRNAs, then added to DNA substrates along with the indicated components. After 1 hour, reactions were analyzed by denaturing PAGE, visualizing Cy5 fluorescence. (e) Primer extension assays performed as in (d) using 3’-extended pegRNAs pre-complexed with dCas9 or Cas9 H840A nickase, and pre-nicked or non-nicked dsDNA substrates. (f) Yeast colonies transformed with GFP–mCherry fusion reporter plasmids edited in vitro with pegRNAs, Cas9 nickase, and RT. Plasmids containing nonsense or frameshift mutations between GFP and mCherry were edited with pegRNAs that restore mCherry translation via transversion, 1-bp insertion, or 1-bp deletion. GFP and mCherry double-positive cells (yellow) reflect successful editing. Images in (d-f) are representative of n=2 independent replicates. For gel source data, see Supplementary Figure 1.