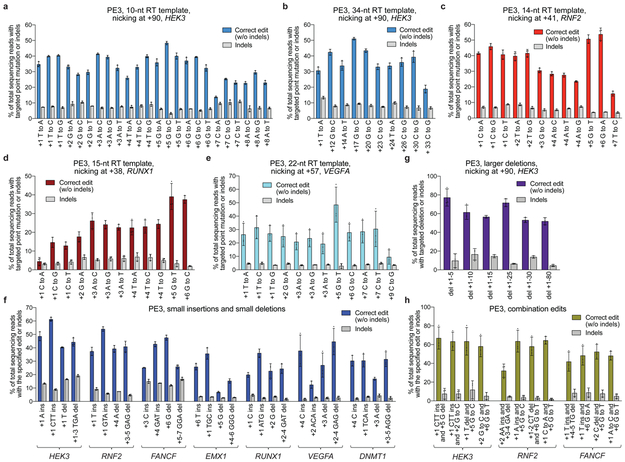
Figure 4. Targeted insertions, deletions, and all 12 types of point mutations with PE3 at seven endogenous genomic loci in HEK293T cells.
(a) All 12 types of single-nucleotide edits from position +1 to +8 of the HEK3 site using a 10-nt RT template, counting the first nucleotide following the pegRNA-induced nick as position +1. (b) Long-range PE3 edits at HEK3 using a 34-nt RT template. (c-e) PE3-mediated transition and transversion edits at the specified positions for (c) RNF2, (d) RUNX1, and (e) VEGFA. (f) Targeted 1- and 3-bp insertions, and 1- and 3-bp deletions with PE3 at seven endogenous genomic loci. (g) Targeted precise deletions of 5-80 bp at HEK3. (h) Combination edits at three endogenous genomic loci. Editing efficiencies reflect sequencing reads that contain the intended edit and do not contain indels among all treated cells, with no sorting. Values and error bars reflect mean±s.d. of n=3 independent biological replicates.