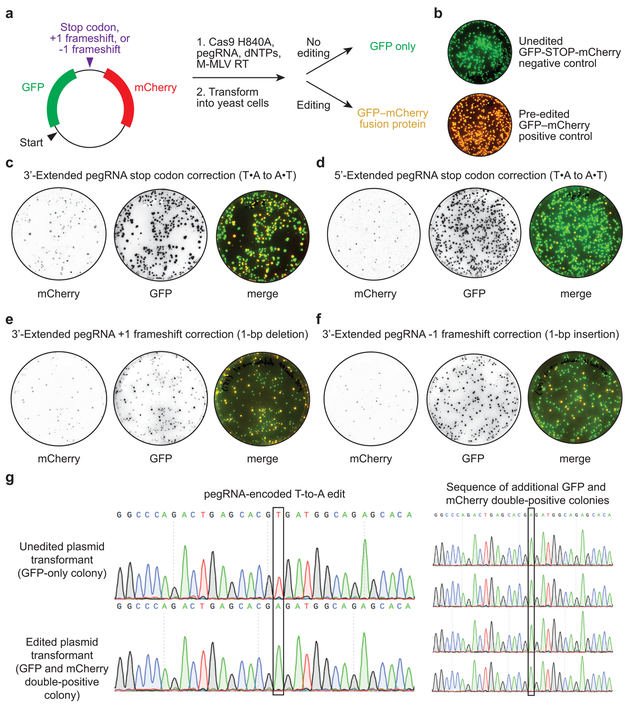
Extended Data Figure 2. Cellular repair in yeast of 3’ DNA flaps from in vitro prime editing reactions.
(a) Dual fluorescent protein reporter plasmids contain GFP and mCherry open reading frames separated by a target site encoding an in-frame stop codon, a +1 frameshift, or a −1 frameshift. Prime editing reactions were carried out in vitro with Cas9 H840A nickase, pegRNA, dNTPs, and M-MLV reverse transcriptase, then transformed into yeast. Colonies that contain unedited plasmids produce GFP but not mCherry. Yeast colonies containing edited plasmids produce both GFP and mCherry as a fusion protein. (b) Overlay of GFP and mCherry fluorescence for yeast colonies transformed with reporter plasmids containing a stop codon between GFP and mCherry (unedited negative control, top), or containing no stop codon or frameshift between GFP and mCherry (pre-edited positive control, bottom). (c-f) Visualization of mCherry and GFP fluorescence from yeast colonies transformed with in vitro prime editing reaction products. (c) Stop codon correction via T•A-to-A•T transversion using a 3’-extended pegRNA or (d) a 5’-extended pegRNA. (e) +1 frameshift correction via a 1-bp deletion using a 3’-extended pegRNA. (f) −1 frameshift correction via a 1-bp insertion using a 3’-extended pegRNA. (g) Sanger DNA sequencing traces from plasmids isolated from GFP-only colonies in (b) and GFP and mCherry double-positive colonies in (c). Data in (b-g) are representative of n=2 independent replicates.