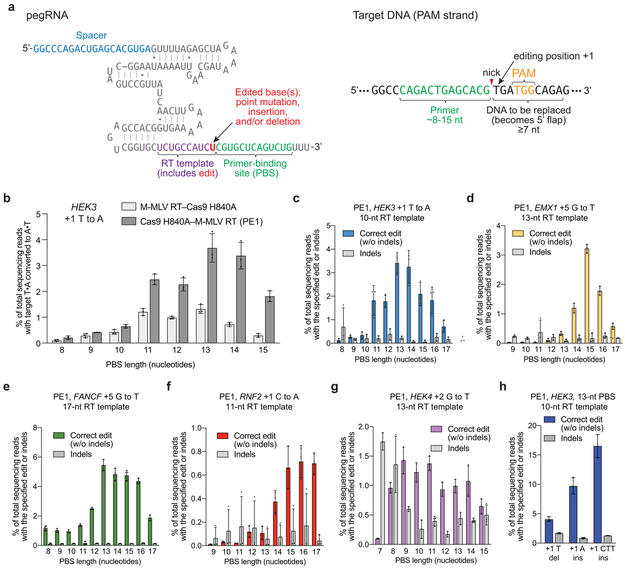
Extended Data Figure 3. Prime editing of genomic DNA in human cells by PE1.
(a) pegRNAs contain a spacer sequence, an sgRNA scaffold, and a 3’ extension containing a reverse transcription (RT) template (purple), which contains the edited base(s) (red), and a primer-binding site (PBS, green). The primer-binding site hybridizes to the nicked target DNA strand. The RT template is homologous to the DNA sequence downstream of the nick, with the exception of the encoded edited base(s). (b) Installation of a T•A-to-A•T transversion at the HEK3 site in HEK293T cells using Cas9 H840A nickase fused to wild-type M-MLV reverse transcriptase (PE1) and pegRNAs with varying PBS lengths. (c) T•A-to-A•T transversion editing efficiency and indel generation by PE1 at the +1 position of HEK3 using pegRNAs containing 10-nt RT templates and a PBS sequences ranging from 8-17 nt. (d) G•C-to-T•A transversion editing efficiency and indel generation by PE1 at the +5 position of EMX1 using pegRNAs containing 13-nt Rt templates and a PBS sequences ranging from 9-17 nt. (e) G•C-to-T•A transversion editing efficiency and indel generation by PE1 at the +5 position of FANCF using pegRNAs containing 17-nt RT templates and a pBs sequences ranging from 8-17 nt. (f) C•G-to-A•T transversion editing efficiency and indel generation by PE1 at the +1 position of RNF2 using pegRNAs containing 11 -nt RT templates and a PBS sequences ranging from 9-17 nt. (g) G•C-to-T•A transversion editing efficiency and indel generation by PE1 at the +2 position of HEK4 using pegRNAs containing 13-nt RT templates and a PBS sequences ranging from 7-15 nt. (h) PE1-mediated +1 T deletion, +1 A insertion, and +1 CTT insertion at the HEK3 site using a 13-nt PBS and 10-nt RT template. Sequences of pegRNAs are those used in Fig. 2a (see Supplementary Table 3). Editing efficiencies reflect sequencing reads that contain the intended edit and do not contain indels among all treated cells, with no sorting. Values and error bars reflect mean±s.d. of n=3 independent biological replicates.