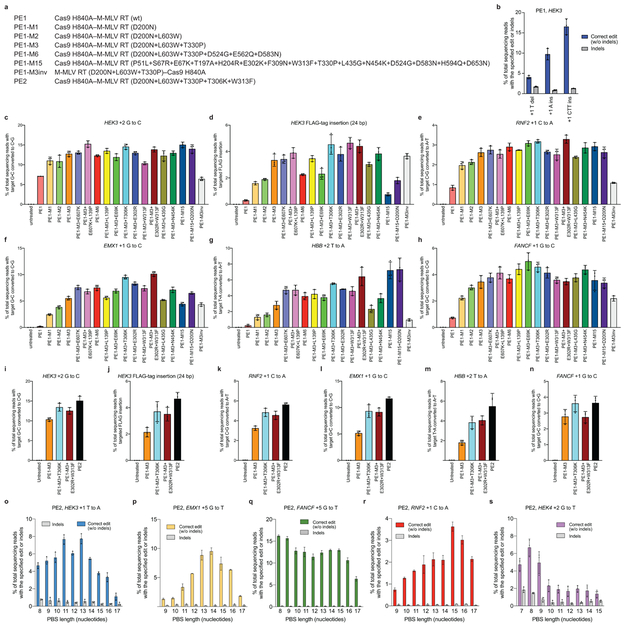
Extended Data Figure 4. Evaluation of M-MLV RT variants for prime editing.
(a) Abbreviations for prime editor variants used in this figure. (b) Targeted insertion and deletion edits with PE1 at the HEK3 locus. (c-h) Comparison of 18 prime editor constructs containing M-MLV RT variants for their ability to install (c) a +2 G•C-to-C•G transversion edit at HEK3, (d) a 24-bp FLAG insertion at the +1 position of HEK3, (e) a +1 C•G-to-A•T transversion edit at RNF2, (f) a +1 G•C-to-C•G transversion edit at EMX1, (g) a +2 T•A-to-A•T transversion edit at HBB, and (h) a +1 G•C-to-C•G transversion edit at FANCF. (i-n) Comparison of four prime editor constructs containing M-MLV variants for their ability to install the edits shown in (c-h) in a second round of independent experiments. (o-s) PE2 editing efficiency at five genomic loci with varying PBS lengths. (o) +1 T•A-to-A•T at HEK3. (p) +5 G•C-to-T•A at EMX1. (q) +5 G•C-to-T•A at FANCF. (r) +1 C•G-to-A•T at RNF2. (s) +2 G•C-to-T•A at HEK4. Editing efficiencies reflect sequencing reads that contain the intended edit and do not contain indels among all treated cells, with no sorting. Values and error bars reflect mean±s.d. of n=3 independent biological replicates.