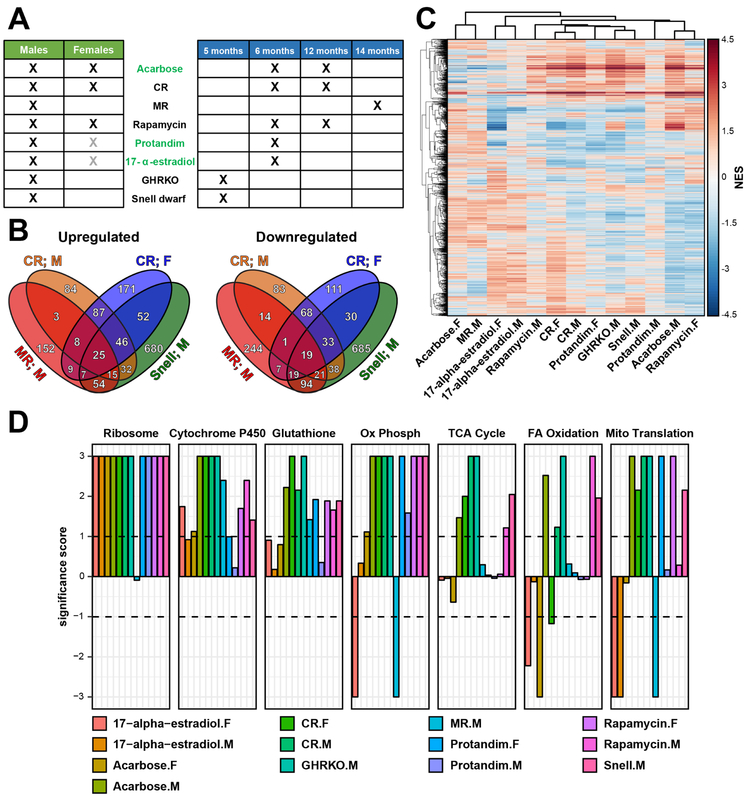
Figure 1. RNAseq of mouse hepatic response to longevity interventions.
(A) RNAseq dataset. X denotes utilized experimental designs (n=3 for each group). Interventions, which have not been previously analyzed at the level of gene expression, are colored in green. Grey X marks denote experimental designs that failed to extend lifespan with statistical significance.
(B) Overlap of significant gene expression changes in response to longevity interventions.
(C) Functions enriched by genes changed in response to lifespan-extending interventions. Normalized enrichment scores (NES) of functions enriched by at least one intervention are shown.
(D) Functions enriched by up- (up) and downregulated (down) genes across interventions. Significance score, calculated as log10(q-value) corrected by the sign of regulation, is plotted on the y axis. FDR threshold of 0.1 is shown by dotted lines. The whole list of enriched functions is in Table S2.
Cytochrome P450: Drug metabolism by cytochrome P450; Glutathione: Glutathione metabolism; Ox Phosph: Oxidative phosphorylation; TCA cycle: Citrate Cycle/TCA Cycle; FA oxidation: Fatty acid β-oxidation; Mito Translation: Mitochondrial translation; Snell: Snell dwarf mice; F: Females; M: Males.