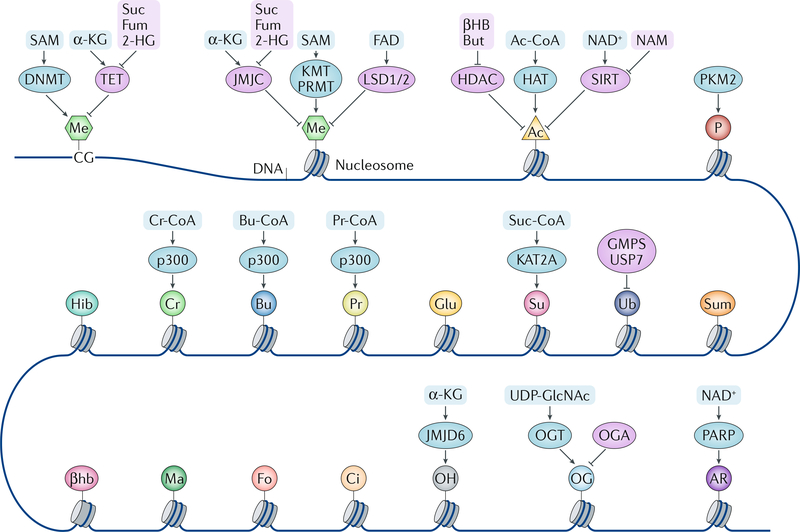
Fig. 1 |. Chromatin modulation by metabolites.
DNA and histones are modified by different writers and erasers, and the enzymatic activities of these modifiers are regulated by metabolites and metabolic enzymes. Some metabolic enzymes can act as direct modifiers (writers) of the histone code as well. α-KG, α-ketoglutarate; βHB, β-hydroxybutyrate; βhb, β-hydroxybutyrylation; 2-HG, 2-hydroxyglutarate; Ac, acetylation; Ac-CoA, acetyl-CoA; AR, ADP ribosylation; Bu, butyrylation; Bu-CoA, butyryl-CoA; But, butyrate; Ci, citrullination; Cr, crotonylation; Cr-CoA, crotonyl-CoA; DNMT, DNA methyltransferase; Fo, formylation; Fum, fumarate; Glu, glutarylation; GMPS, GMP synthase; HAT, histone acetyltransferase; HDAC, histone deacetylase; Hib, 2-hydroxyisobutyrylation; JMJC, Jumonji C domain-containing demethylase; JMJD6, Jumonji domain-containing 6; KMT, lysine methyltransferase; LSD, lysine-specific histone demethylase; Ma, malonylation; Me, methylation; NAM, nicotinamide; OG, O-GlcNAcylation; OGA, O-GlcNAcase; OGT, O-GlcNAc transferase; OH, hydroxylation; P, phosphorylation; PARP, poly(ADP) ribose polymerase; PKM2, pyruvate kinase M2 isoform; Pr, propionylation; Pr-CoA, propionyl-CoA; PRMT, peptidyl-arginine methyltransferase; SAM, S-adenosylmethionine; SIRT, sirtuin; Su, succinylation; Suc, succinate; Suc-CoA, succinyl-CoA; Sum, sumoylation; TET, ten-eleven translocation (DNA demethylase); Ub, ubiquitylation; UDP-GlcNAc, uridine diphosphate N-acetylglucosamine; USP7, ubiquitin-specific processing protease 7.