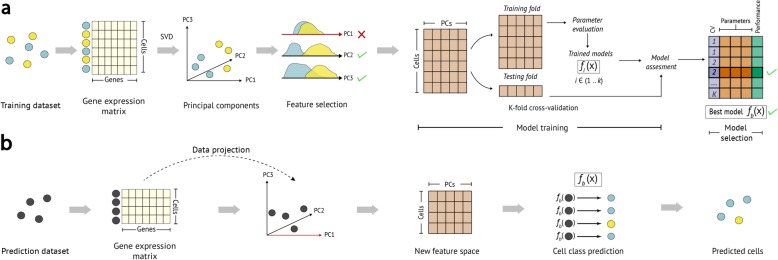
Fig. 1.
Summary of the scPred method. a Training step. A gene expression matrix is eigendecomposed via singular value decomposition (SVD) to obtain orthonormal linear combinations of the gene expression values. Only PCs explaining greater than 0.01% of the variance of the dataset are considered for the feature selection and model training steps. Informative PCs are selected using a two-tailed Wilcoxon signed-rank test for each cell class distribution (see the “Methods” section). The cells-PCs matrix is randomly split into k groups and the first k group is considered as a testing dataset for cross-validation. The remaining K-1 groups (shown as a single training fold) are used to train a machine learning classification model (a support vector machine). The model parameters are tuned, and each k group is used as a testing dataset to evaluate the prediction performance of a fi(x) model trained with the remaining K-1 groups. The best model in terms of prediction performance is selected. b Prediction step. The gene expression values of the cells from an independent test or validation dataset are projected onto the principal component basis from the training model, and the informative PCs are used to predict the class probabilities of each cell using the trained prediction model(s) fb(x)