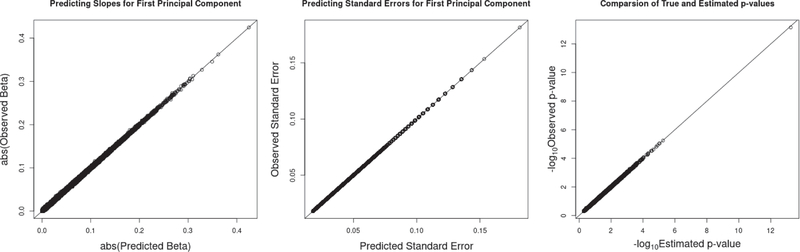
Fig. 1:
Differences of our method’s approximations of slope, standard error of slope, and p-values and those achieved when fitting a model for the first principal component on the raw data. These figures illustrate the high accuracy of our method, even when approximating the covariance structure of the phenotypes.
(a) Difference of observed and predicted SNP slope coefficients on simulated data when approximating phenotype covariance.
(b) Difference of observed and predicted standard errors of the SNP slope coefficient on simulated data when approximating phenotype covariance.
(c) Difference of observed and predicted p-values of SNPs and the first principal component on simulated data when approximating phenotype covariance.(−log10 scale)