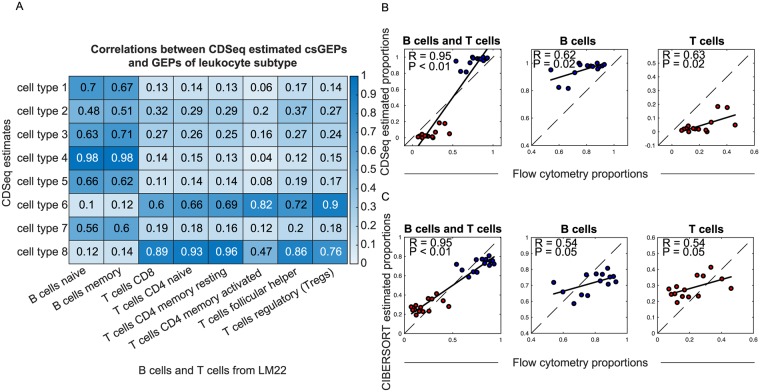
Fig 8. Comparison of CDSeq using the quasi-unsupervised strategy with CIBERSOFT on deconvolution of B cells and T cells in lymphoma samples.
We ran CDSeq with 22 cell types, α = 0.5, β = 0.5, and 700 MCMC runs. We considered an anonymous CDSeq-identified cell type to match one of the B cell (blue dots) or T cell subtypes (red dots) if the Pearson correlation of their GEPs exceeded 0.6. (A) Correlation between estimated GEPs and true GEPs; (B) CDSeq estimated proportions versus flow cytometry; (C) CIBERSORT estimation versus flow cytometry.