FIGURE 2. Power of unified likelihood and regression imputation strategies for normalized expression data.
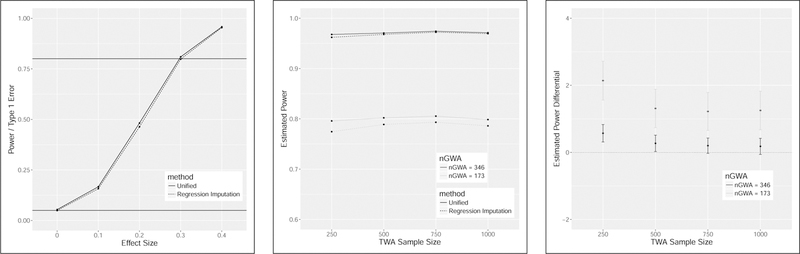
Power and type-1 error rates in the left-hand panel are estimated based 10,000 simulations per condition and a significance threshold of 0.05. The black horizontal lines have y-intercepts of 0.05 and 0.80. For the left hand panel, the observed data sample sizes of nGWA=173 and nTWA=906 are assumed and a range of effect sizes (μ/oc ) are considered. For the center and right hand panel an expression effect size of 0.3 is assumed and the observed genotype data are used. Assumed parameter values are based on application to the CARMIL1, previously LRRC16A, gene (Table 5). A range of nTWA sample sizes are considered and the sample size of nGWA is assumed to be the observed sample size and then inflated by a factor of 2. Power remains relatively constant for nTWA sample sizes ranging from 250 to 1000. Power increases substantially with larger GWA sample size. Power of the likelihood-based approach is consistently higher than the regression imputation approach, though the difference is between 0 and 2% for the scenarios considered. This difference is statistically significant for the observed sample sizes.
