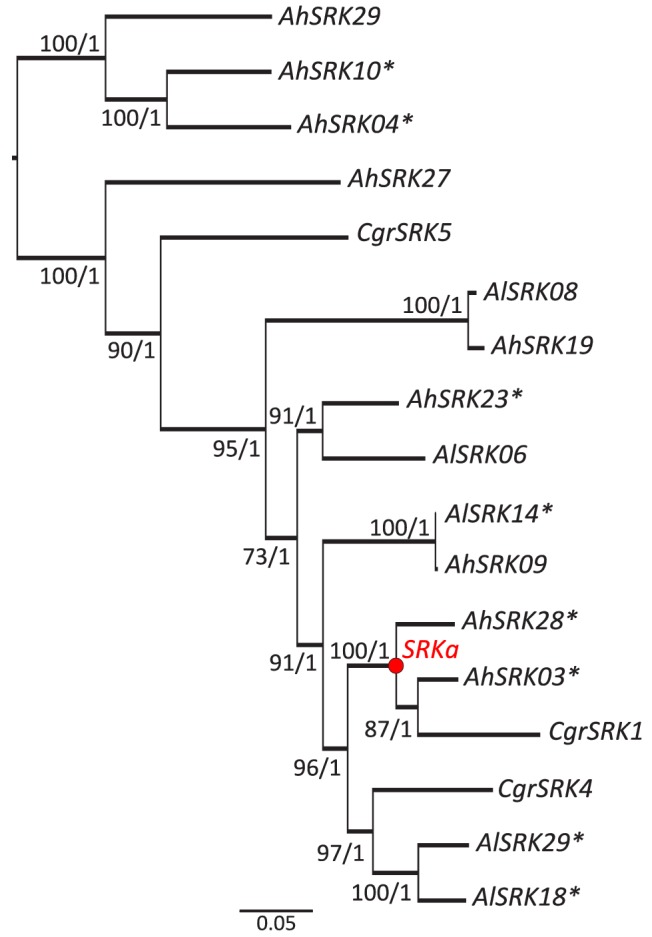
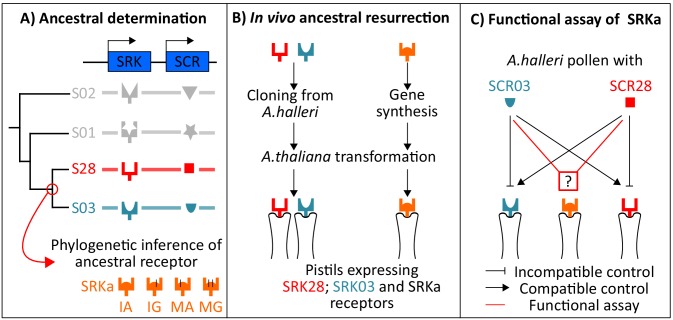
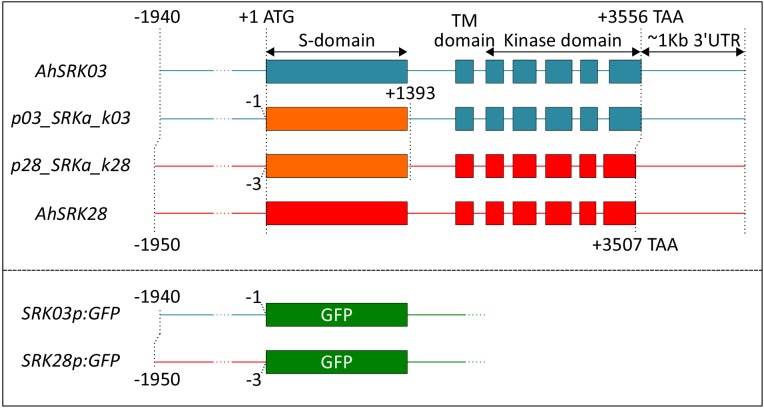
Figure 1. Experimental approach for the ancestral resurrection experiment.
(A) The sequence of the putative last common ancestor of SRK03 and SRK28 was inferred by a phylogenetic approach using codon-based models implemented in PAML. Four different versions of SRKa were defined due to inference uncertainty at two aa positions. (B) SRK03 and SRK28 sequences were cloned from A. halleri DNA BAC clones, whereas SRKa sequences were obtained by gene synthesis. (C) Representation of the controlled cross program to decipher the specificity of SRKa.
Figure 1—figure supplement 1. Three models for the emergence of new self-incompatibility specificities.
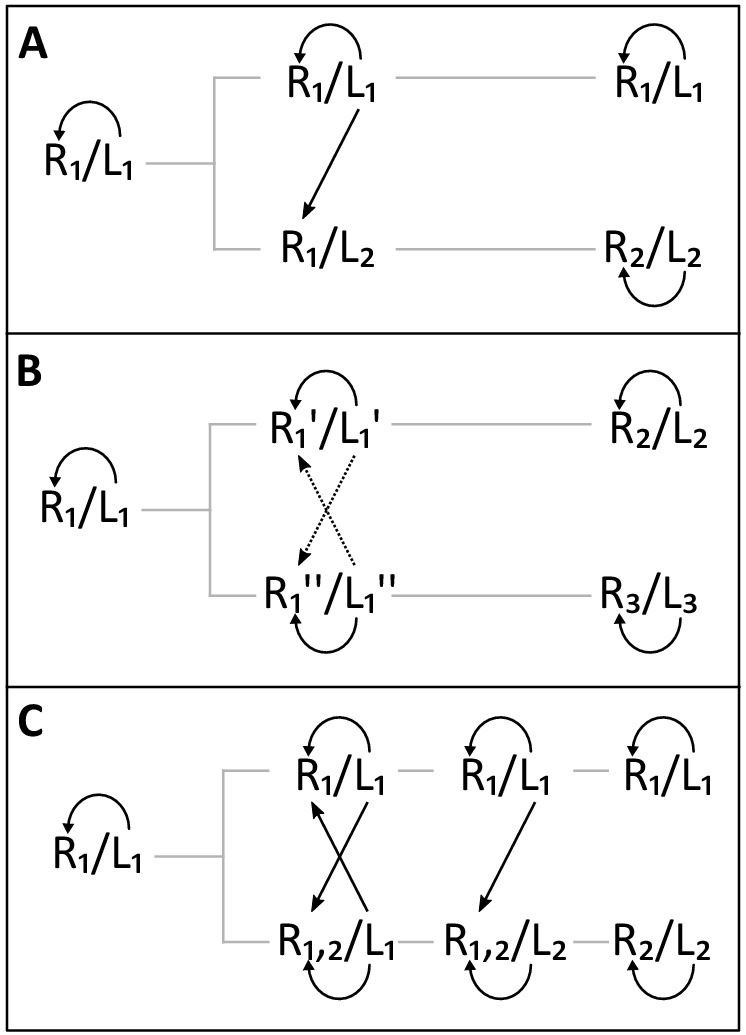
Figure 1—figure supplement 2. Maximum likelihood phylogenetic tree based on the 17 SRK alleles used for SRKa construction.