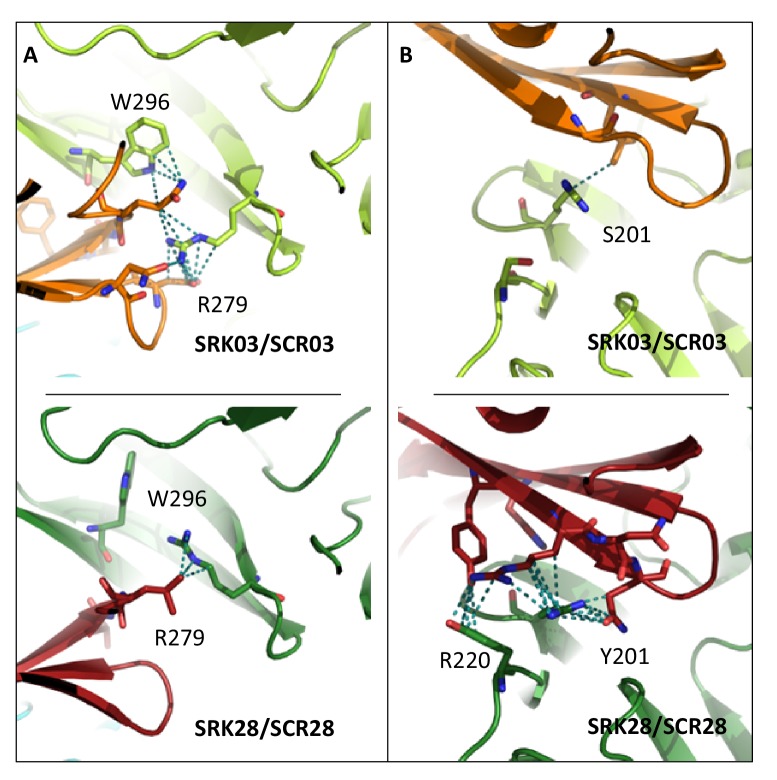
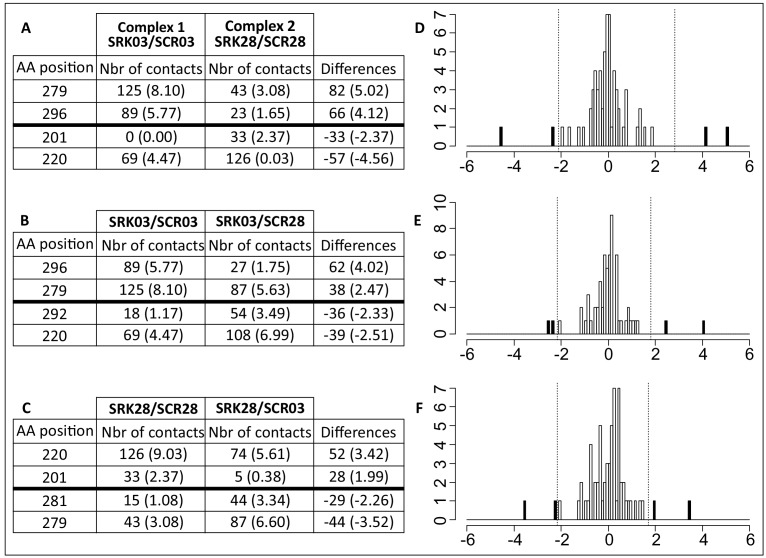
Figure 5. Four amino acid residues establish contrasted patterns of atomic contacts between SCR and SRK in the two cognate complexes.
In (A), residues R279 and W296 of eSRK establish a large number of atomic contacts with SCR in the S03 complex (upper panel), but a very low number of contacts in the S28 complex (lower panel). The situation is reversed in (B), where residues S201 and R220 of eSRK establish a large number of atomic contacts with SCR in the S28 complex (lower panel), but a very low number of contacts in the S03 complex (upper panel). SRK chains are coloured in light and dark green for SRK03 and SRK28, respectively; SCR chains are coloured in orange and red for SCR03 and SCR28, respectively. Amino acid residues are shown in stick representation, with dotted lines indicating atom pair contacts below 4 Å, excluding hydrogen atoms. Note that for clarity a more stringent threshold was used to define atomic contacts here (4 Å) than in Figure 5—figure supplement 1 and Figure 6—figure supplement 1 (where a 5 Å threshold was used for a more comprehensive analysis), but the results are qualitatively similar.