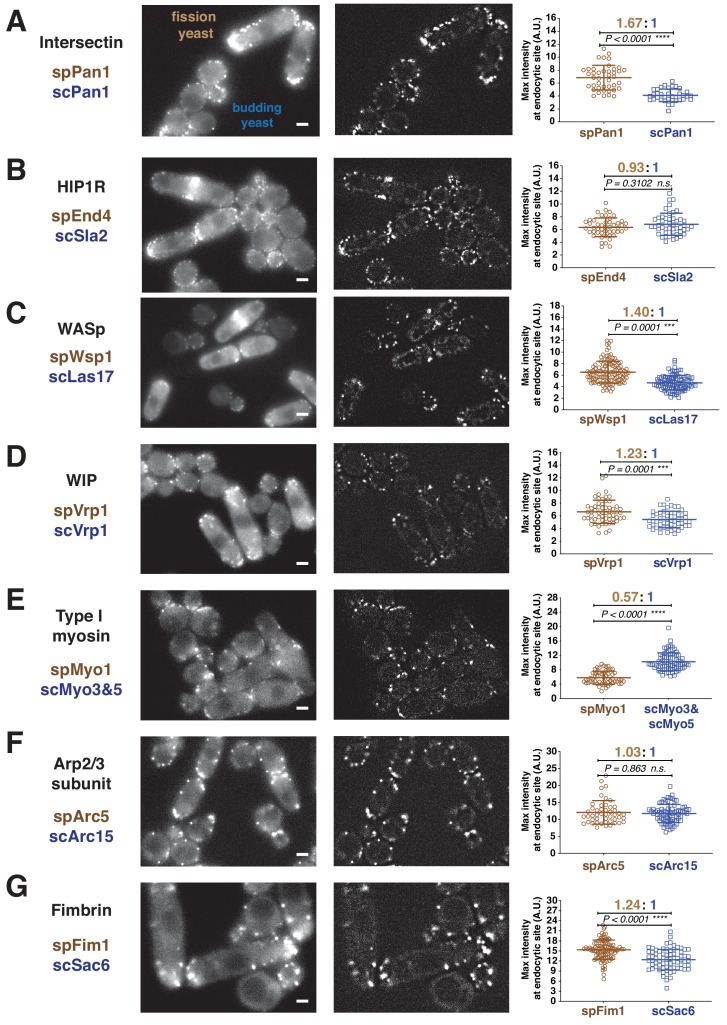
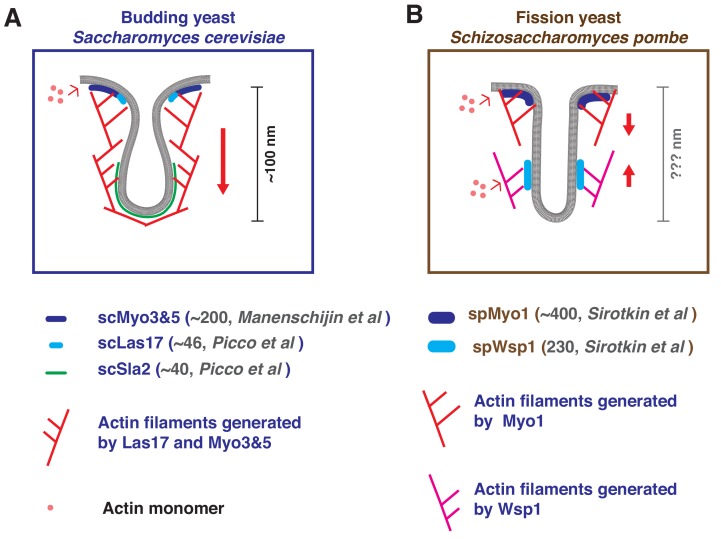
The endocytic actin machinery on invaginated membranes in budding (
A) or fission (
B) yeast. (
A) In budding yeast, actin filaments are proposed to be nucleated near the base of the invagination. Continued polymerization driven by the nucleation promoting activity (NPF) of scMyo3/5 and scLas17 pushes the actin network toward cytoplasm (
Sun et al., 2006). The endocytic membrane is coupled to the actin network by coat proteins such as Sla2 and is pulled inward with the actin network (
Kaksonen et al., 2003;
Sun et al., 2005). The membrane invagination is ~100 nm deep at the time of vesicle scission (
Kukulski et al., 2012). Previously, HA-tagged Myo5 was observed not only at the base but also at the tip of endocytic membrane invaginations in chemically fixed budding yeast cells by immunoelectron microscopy (
Idrissi et al., 2008;
Idrissi et al., 2012). These results led to the proposal that two distinct endocytic actin networks exist in budding yeast. However, two independent live-cell imaging studies indicated that Myo5-GFP patches stay non-motile at the cell cortex during their lifetime (
Galletta et al., 2008;
Sun et al., 2006). Furthermore, super-resolution microscopy suggested that Myo5-GFP forms a Nano-template for actin nucleation at the membrane base (
Mund et al., 2018). Together, these observations suggest that most if not all of the type I myosin is localized at the base of invaginated endocytic membranes. (
B) In fission yeast, a two-zone model proposes that actin nucleation by spMyo1 and spWsp1generate two independent actin networks that push against each other and pull the tip of the invagination into the cytoplasm (
Arasada and Pollard, 2011). The depth of the fission yeast membrane invagination is not clear. Note, some of the previously measured absolute molecular numbers of indicated protein homologues differ greatly between the two yeasts (
Manenschijn et al., 2019;
Picco et al., 2015;
Sirotkin et al., 2010).