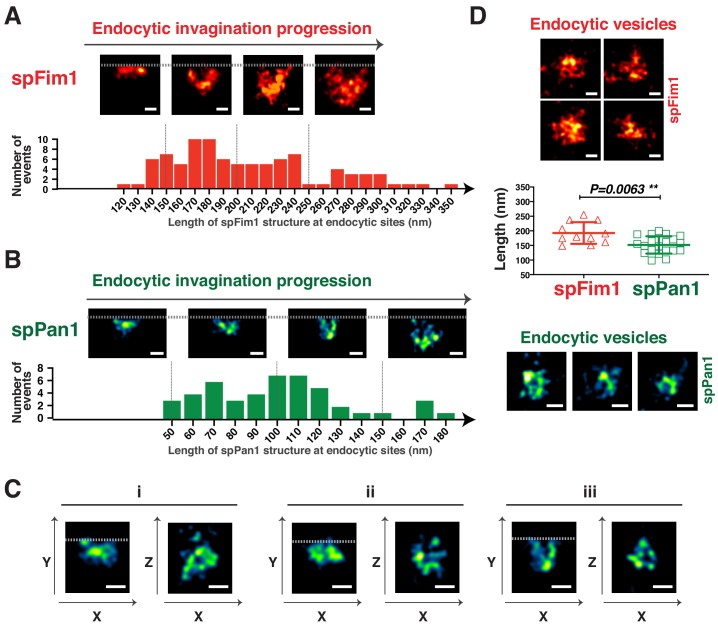
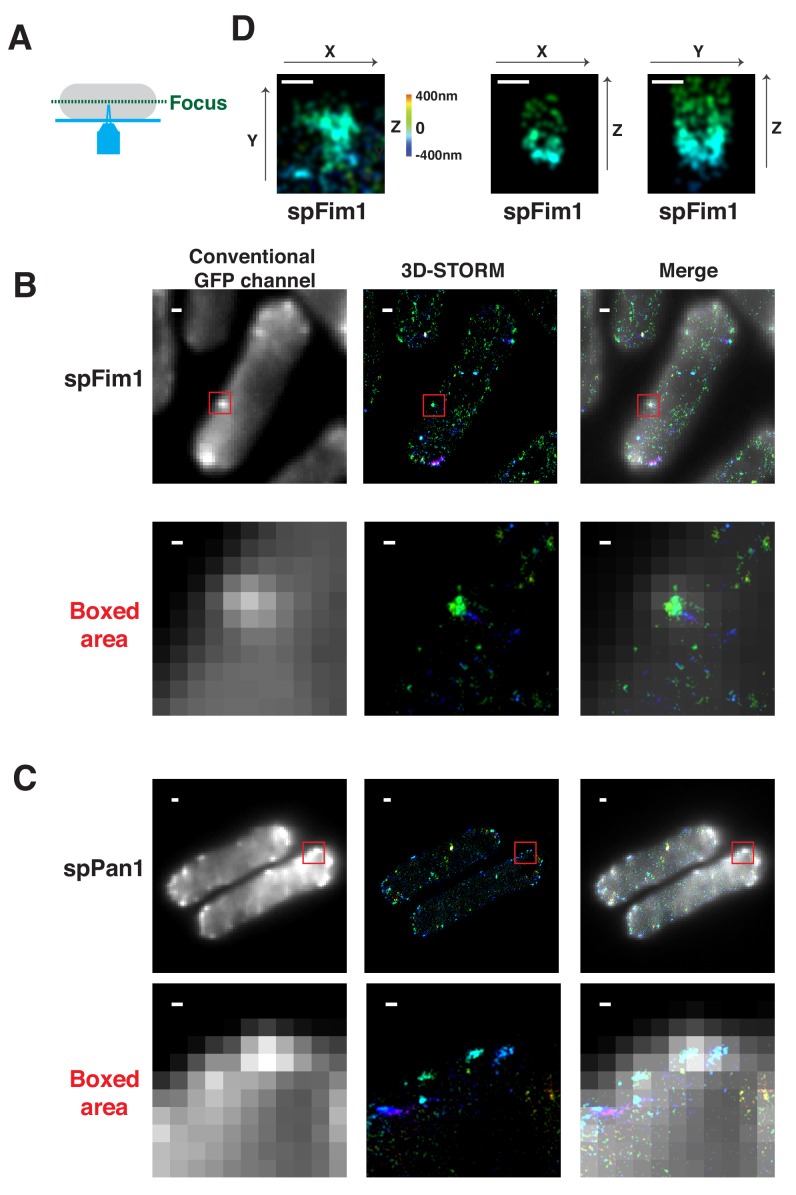
Figure 6. 3D-STORM imaging of coat protein and the actin network at fission yeast endocytic sites.
(A and B) The length of nanoscale structures of spFim1 (n = 92) or spPan1 (n = 46) at endocytic sites revealed by STORM analysis was measured and plotted. Representative STORM images of different length and quantitative analysis of spFim1 (A) and spPan1 (B). The histogram shows the frequency distribution of observed endocytic structure lengths. C, 3D-STORM image analysis reveals ring-like Pan1 organization in the XZ dimensions. D, Representative images of presumed endocytic vesicles in the cytoplasm. The length of longest axis of each presumed vesicle was measured and plotted. Fim1-labeled structures were pseudo-colored red hot (A and D) and Pan1-labeled structures were pseudo-colored green fire blue (B, C, D). The scale bars on STORM images are 100 nm.