FIGURE 1.
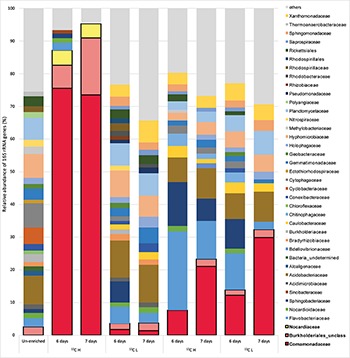
Relative abundance of 16S rRNA genes at the family level in DNA isolated after DNA-SIP enrichments. The relative abundance of bacterial 16S rRNA genes retrieved by PCR from un-enriched (unfractionated) DNA, extracted after sampling, is shown on the left. Subsequent bars show the relative abundance of 16S rRNA genes retrieved from DNA-SIP experiments with isoprene-enriched willow soil samples after 6 and 7 days of enrichment with 12C- and 13C-labeled isoprene. DNA arising from willow soil samples enriched with 12C-isoprene and 13C-isoprene are designated as light (L) and heavy (H) fractions respectively (refer to Supplementary Figures S5, S6). Taxonomic affiliation of 16S rRNA genes is reported at the family level. Only 16S rRNA gene sequences with a relative abundance of greater than 1% are shown. 16S rRNA gene sequences with a relative abundance of less than 1% are grouped together as “others.” Families (Comamonadaceae, Burkholderiales unclassified, and Nocardiaceae) identified in these DNA-SIP experiments as putative isoprene-degrading bacteria according to 13C-labeling, are highlighted with a black border.
