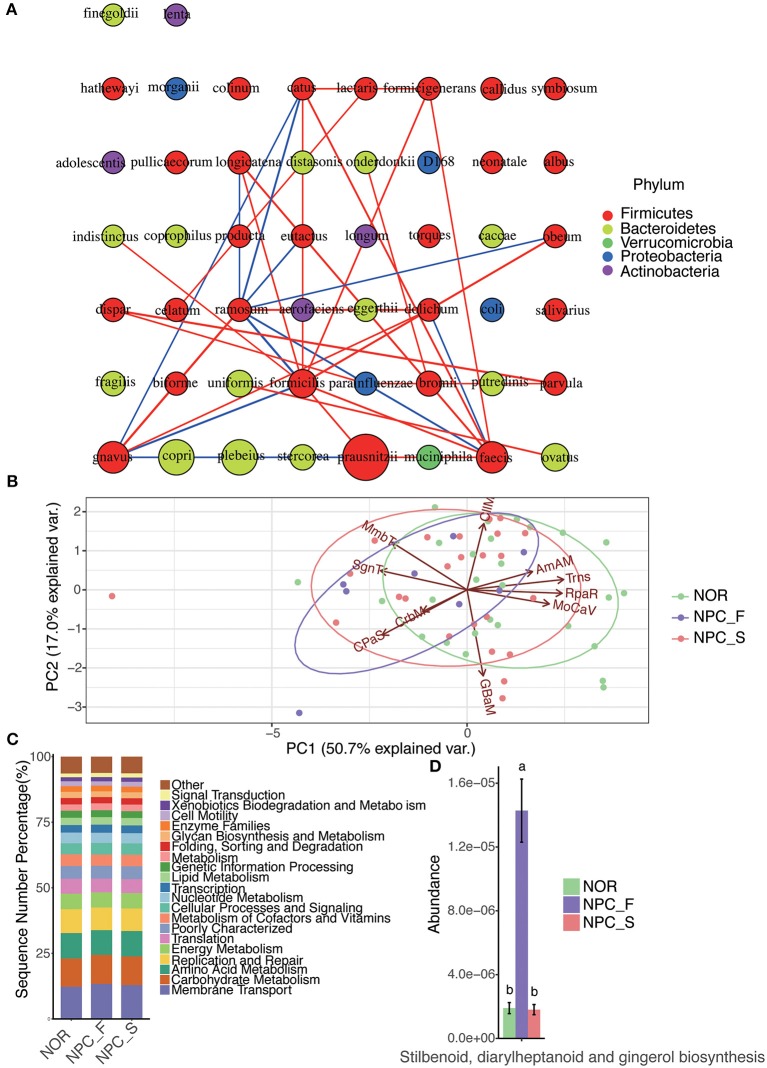
Figure 13.
The intestinal flora interaction network analysis and functional PICRUSt analysis among the healthy controls (NOR), familial NPC patients (NPC_F) and sporadic NPC patients (NPC_S) groups. (A) The map of the species interaction network analysis; a circle represents a bacteria, the size of the circle represents its relative abundance, different color represents different classification at phylum level, the line between the circles represents the correlation between the two bacteria is significant (P < 0.05), the red color of the line represents a positive correlation, while the blue one represents the negative correlation, the line is more rough, corresponding correlation coefficient value is greater. (B) The diagram of PCA analysis for the predicted functions on the second KEGG-pathway level; The PC1-axis (50.7%) and PC2-axis (17.0%) represent the contribution of the two principal components to the sample difference are 50.7% and 17.0%, respectively, each point represents a sample, each circle represents a group (NOR in green, NPC_F in purple and NPC_S in red), and the arrow direction and length represent the direction and dominant ability of the prediction function in the group, respectively. (AmAM, Amino Acid Metabolism; CIIM, Cell Motility; CPaS, Cellular Processes and Signaling; CrbM, Carbohydrate Metabolism; GBaM, Glycan Biosynthesis and Metabolism; MmbT, Membrane Transport; MoCaV, Metabolism of Cofactors and Vitamins; RpaR, Replication and Repair; SgnT, Signal Transduction; Trns, Translation). (C) The bar chart of the predicted functions at the second KEGG-pathway level. (D) The bar chart of the abundance of the function of stilbenoid, diarylheptanoid and gingerol biosynthesis of the NOR, NPC_F and NPC_S groups, the value of a and b represent that there is significant difference in abundance of this function compared NPC_F with NOR, and NPC_F with NPC_S groups.