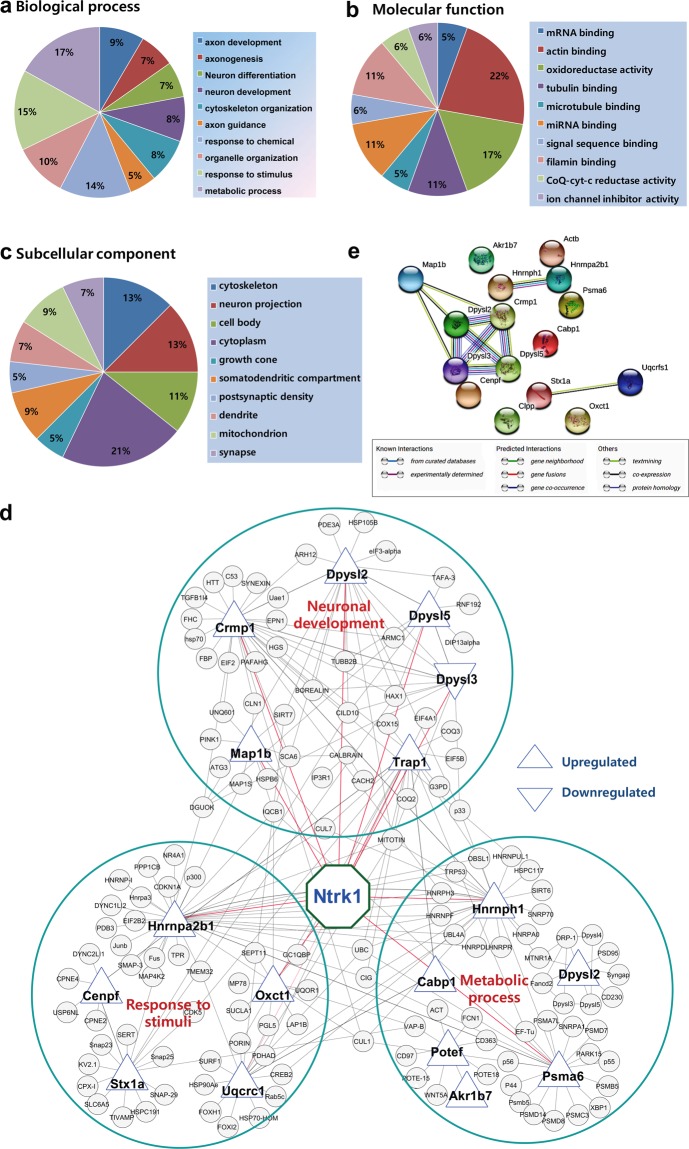
Figure 5.
Bioinformatics analysis of the differentially expressed proteins. Functional enrichment of Gene Ontology (GO) by DAVID annotation tool (version 6.8): top 10 GO terms for biological process (a), molecular function (b) and subcellular component (c) were displayed. Interaction networks were visualized by Cytoscape software (version 3.7.1) (d). Up and down triangles indicate up- and down-regulated proteins, respectively. Large grouping circles indicate three enriched protein−protein interaction clusters. Here interaction of TrkA (Ntrk1) with the majority of the differentially regulated proteins indicates that SCH might function through TrkA signaling pathway. Protein-protein interaction network analysis by STRING (version 11.0) (e).