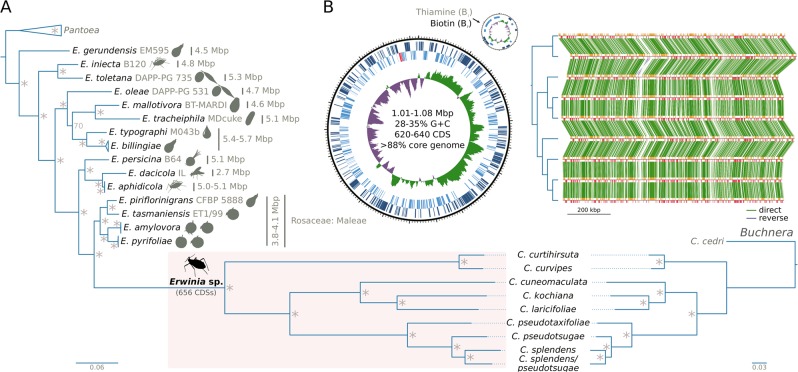
Fig. 2.
Phylogenetic reconstruction of Erwinia spp. and genome properties of Cinara-associated Erwinia endosymbionts. a On the left, phylogenetic reconstruction for Erwinia spp., using Pantoea spp. as an outgroup. Erwinia endosymbionts of Cinara species form a well-supported monophyletic group. Silhouettes next to the leaves represents the isolation source for the strains. For the Erwinia symbionts, the species name of the Cinara host is used. On the bottom right, the phylogeny of the corresponding Buchnera strains, using the Buchnera associated with C. cedri as an outgroup, is shown. Asterisks at nodes of trees stand for a posterior probability equal to 1. b On the left, the genome plot for the endosymbiont of C. pseudotaxifoliae along with statistics for sequenced Erwinia genomes. From outermost to innermost ring, the features on the direct strand, the reverse strand, and GC-skew plot are shown. On the right, pairwise synteny plots of Erwinia endosymbionts with a dendogram on the left displaying their phylogenetic relationships