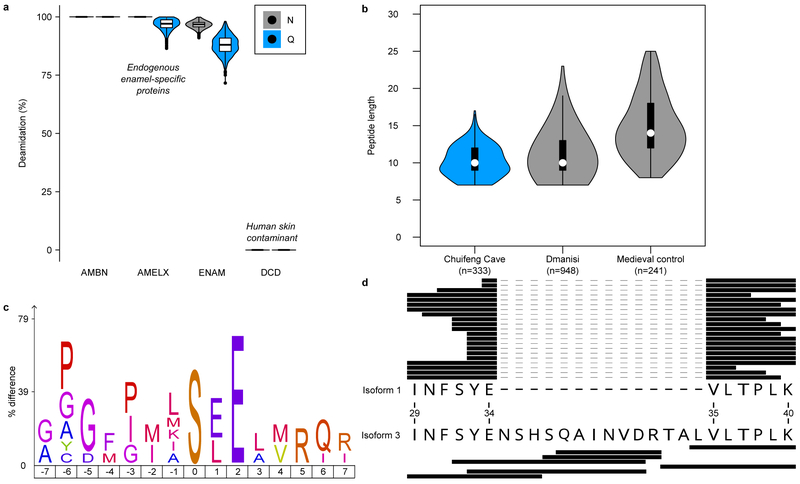
Figure 2. Gigantopithecus enamel proteome modifications and degradation.
a, Violin plots of asparagine (N) and glutamine (Q) deamidation for selected proteins (n=1,000 bootstrap replicates of intensity-based peptide deamidation32). Human dermcidin (DCD) is included as an example of a non-deamidated contaminant. For AMBN, all observed asparagines and glutamines are deamidated. For AMELX, all asparagines are deamidated. For DCD, no observed asparagines and glutamines are deamidated. b, Violin plots of peptide lengths for Gigantopithecus, an Early Pleistocene rhinoceros from Dmanisi, and a Medieval control sample17. c, Sequence-motif analysis of the over-representation of specific amino acids around the phosphorylated amino acid (position 0; n=14). d, Peptide coverage of AMELX protein isoforms. Matching peptides are indicated by black bars for isoform 1 (n=21) and isoform 3 (n=7). The latter includes an insertion due to alternative splicing between isoform 1, amino acid positions 34 and 35 (coordinates in reference to UniProt Accession number: Q99217-1 [AMELX_HUMAN]). a and b include data on AMELX, AMBN, ENAM, AMTN and MMP20 only. For a and b, boxplots define the range of the data (whiskers extending to 1.5 the interquartile range), 25th and 75th percentiles (boxes), and medians (dots).