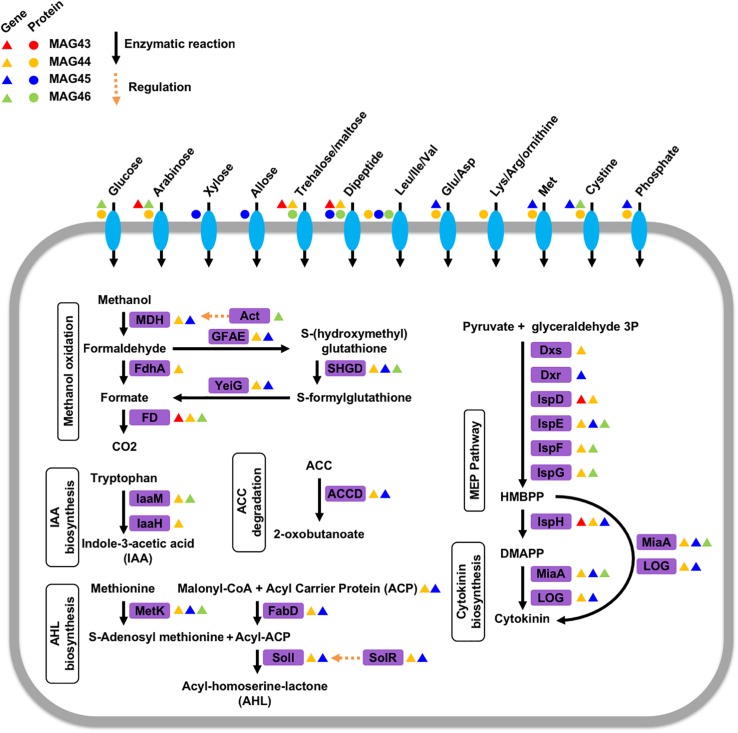
FIGURE 2.
Phytohormone modulation, nutrient transportation, quorum sensing, and methanol metabolism in the four 13C-labeled microorganisms. Enzymes in purple boxes are labeled with triangles for encoded genes and circles for expressed proteins in these microorganisms. The triangles and circles are color-coded for each microorganism. MDH: NAD-dependent methanol dehydrogenase, Act: methanol dehydrogenase activator, FdhA: glutathione-independent formaldehyde dehydrogenase, FD: formate dehydrogenase, GFAE: glutathione-dependent formaldehyde-activating enzyme, SHGD: S-(hydroxymethyl)glutathione dehydrogenase, YeiG: S-formylglutathione hydrolase, laaM: tryptophan 2-monooxygenase, laaH: indole acetamide hydrolase, MetK: methionine adenosyltransferase, ACCD: 1-aminocyclopropane-1-carboxylate deaminase, FabD: malonyl CoA-acyl carrier protein transacylase, Soil: acyl-homoserine-lactone synthase, SoIR: transcriptional activator/quorum-sensing receptor, Dxs:1-deoxy-D-xylulose 5-phosphate synthase, Dxr: 2-C-methyl-D-erythritol 4-phosphate synthase, IspD: 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, IspE: 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase, IspF: 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, IspG: 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate synthase, IspH: 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, MiaA: tRNA isopentenyltransferase, LOG: cytokinin-specific phosphoribohydrolase ‘Lonely guy’, HMBPP: (E)-4-Hydroxy-3-methyl-but-2-enyl pyrophosphate, DMAPP: dimethylallyl pyrophosphate.