FIGURE 3.
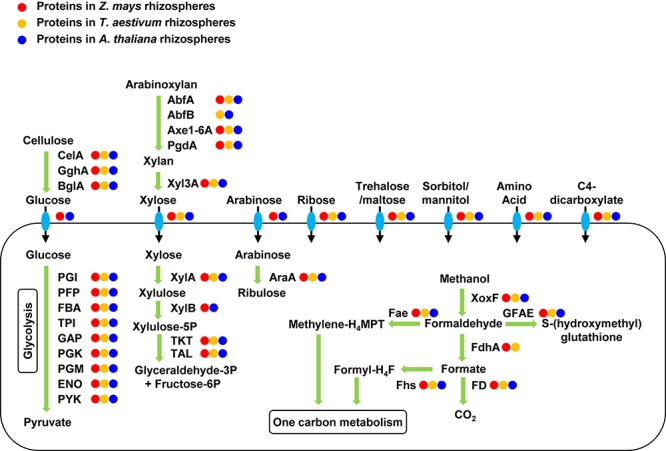
Expressed proteins for carbon uptake and metabolism in the rhizosphere communities. Enzymes are labeled with circles in three different colors for their protein expression in the rhizospheres of the three different plants. The rhizosphere communities expressed proteins for the transportation and metabolism of many complex carbohydrates and methanol. CelA, endo-1,4-beta-D-glucanase; GghA, exo-1,4-beta-glucosidase; BgIA, beta-glucosidase; PGI, phosphoglucose isomerase; PFP, pyrophosphate-dependent fructose-6P 1-phosphotransferase; FBA, fructose-bisphosphate aldolase; TPI, triose-phosphate isomerase; GAP, glyceraldehyde-3P dehydrogenase; PGK, phosphoglycerate kinase; PGM, phosphoglycerate mutase; ENO, enolase; PYK, pyruvate kinase; AbfA, alpha-arabinosidase; AbfB, alpha-arabinosidase; Axe1-6A, feruloyl esterase; PgdA, acetylxylan esterase; Xyl3A, xylan 1,4-beta-xylosidase; XylA, xylose isomerase; XylB, xylulose kinase; TKT, transketolase; TAL, transaldolase; AraA, arabinose isomerase; XoxF, methanol dehydrogenase; FdhA, glutathione-independent formaldehyde dehydrogenase; FD, formate dehydrogenase; GFAE, glutathione-dependent formaldehyde-activating enzyme, Fae, 5,6,7,8-tetrahydromethanopterin hydro-lyase; Fhs, formate-tetrahydrofolate ligase.
