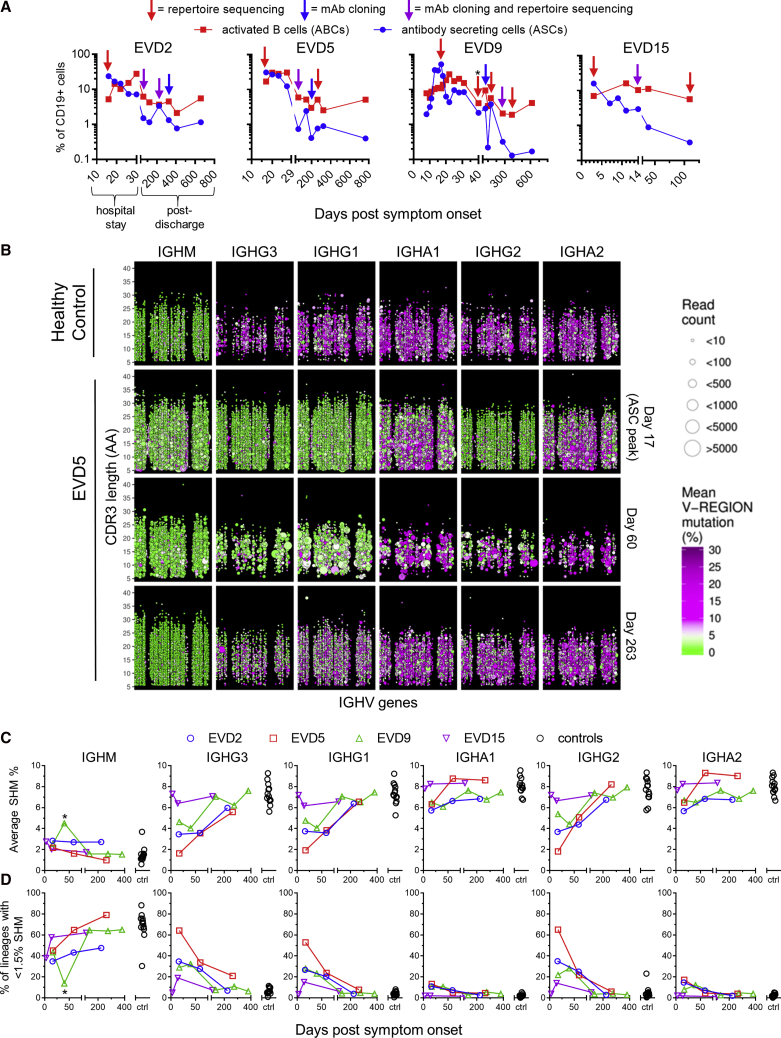
Figure 2.
Profiling of Antibody Sequences in EVD Survivors
(A) Frequencies of activated B cells (ABCs) and antibody secreting cells (ASCs). ABCs were defined as CD20+ CD19+ CD71hi cells and ASCs as CD19+ CD38hi CD27hi CD20 cells. Arrows indicate samples used for repertoire sequencing or monoclonal antibody (mAb) production. The leftmost arrow on each plot corresponds to the ASC peak. Total PBMC RNA was used for repertoire sequencing except for EVD9 day 40 (∗), where RNA from magnetically sorted ASCs was used.
(B) Overview of antibody repertoire in EVD5. The IGH repertoires of a control subject (row 1) and EVD5 (rows 2–4) are shown. Each point represents a single antibody lineage. Point sizes are proportional to the number of unique reads in each lineage, positions indicate the IGHV usage and CDR3 length, and colors indicate the mean somatic hypermutation (SHM). Points are jittered to prevent over-plotting of lineages with the same V and CDR3 lengths. Time points are in days post symptom onset.
(C and D) Summary of somatic hypermutation levels in repertoire sequencing. The average SHM levels of all lineages (C) and the percentage of “low mutation” lineages with SHM <1.5% (D) are compared with 13 control subjects. ∗The second time point for EVD9 used sorted ASC RNA.
See also Figure S2.