Figure 6.
Proposed Self-Resistance Mechanism for the Biosynthesis of MIN
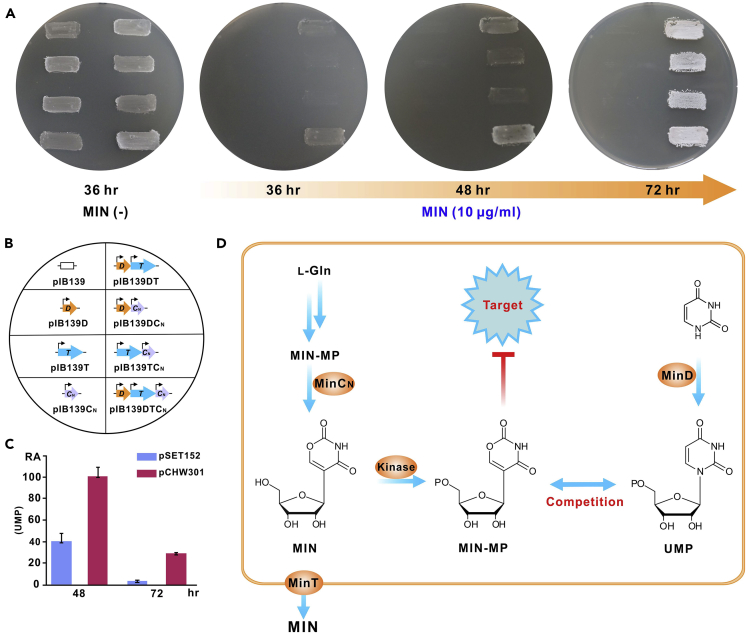
(A) Plate-grown experiments for S. coelicolor M1154 recombinants containing related genes. For the first two plates, the recombinant incubated for 36 h (in the related square) corresponds to each other. For plates 2–4, it means the identical plate that was incubated for different time (marker at the bottom of each plate); MIN (-), the MIN-negative plate; MIN (10 μg/mL), the plate containing MIN at a final concentation of 10 μg/mL. The recombinant in the related square corresponds to that described as below.
(B) Representational map for the plate-grown experiments. pIB139, S. coelicolor M1154 containing the empty vector pIB139; pIB139D, S. coelicolor M1154 containing pIB139/minD; pIB139T, S. coelicolor M1154 containing pIB139/minT; pIB139CN, S. coelicolor M1154 containing pIB139/minCN; pIB139DT, S. coelicolor M1154 containing pIB139/minDT; pIB139DCN, S. coelicolor M1154 containing pIB139/minDCN; pIB139TCN, S. coelicolor M1154 containing pIB139/minTCN; pIB139DTCN, S. coelicolor M1154 containing pIB139/minDTCN.
(C) The relative abundance of the UMP concentrations in vivo for the strains at different growth stages (48 and 72 h). pSET152, the sample of S. coelicolor M1154::pSET152 (blue); pCHW301, the sample of S. coelicolor M1154::pCHW301 (red); “RA” denotes relative abundance. Data are represented as mean ± SEM.
(D) Proposed collaborative self-resistance system during MIN biosynthesis. Three enzymes, including MinCN, MinT, and MinD, collaborate to fulfill the mission of self-resistance during MIN biosynthesis, and MinD, acting as the key safeguard enzyme, employs an unprecedented strategy of substrate competition to achieve self-resistance during MIN biosynthesis.