Figure 2.
Inhibitory Activity against mRNA Processing of Apigenin and Luteolin
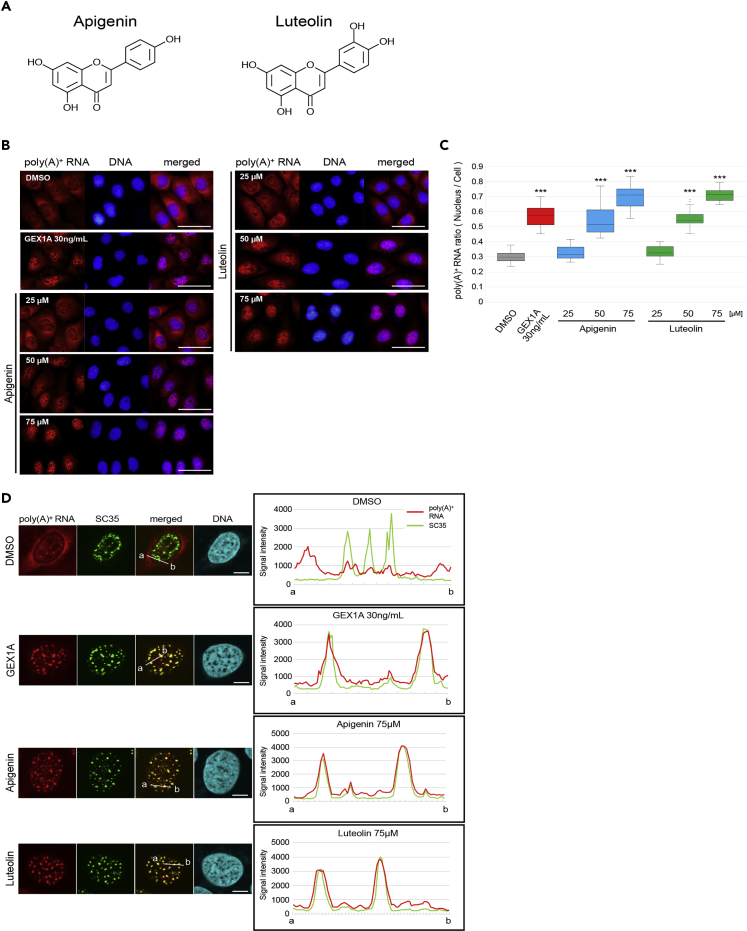
(A) Chemical structures of apigenin and luteolin.
(B) RNA-FISH was performed to determine the localization of bulk poly(A)+ RNA. U2OS cells were treated with the indicated concentrations of each compound for 24 h GEX1A (30 ng/mL) was used as a positive control. The bulk poly(A)+ RNA was visualized by Alexa Fluor 594-labeled oligo-dT45 probe. The nuclei were visualized with DAPI. Scale bar, 50 μm.
(C) The ratio of the nuclear distribution of mRNA was analyzed. The signal intensities of the whole cell and the nucleus were quantified using ImageJ (n = 35). Boxes show median (center line) and upper and lower quartiles. Whiskers show the lowest and highest values. Statistical analysis was performed using one-way ANOVA followed by Dunnett's test. ***p < 0.001.
(D) The localization of bulk poly(A)+ RNA and nuclear speckles. Poly(A)+ RNA (red), speckles (green), and chromosomal DNA (blue) were visualized in U2OS cells for co-localization analysis. Cells were treated with each indicated compound for 24 h. Scale bar, 10 μm. In right panels, signal intensities of poly(A)+ RNA and SC35 were plotted between a and b lines in the left panels. Poly(A)+ RNA and SC35 signal are shown with red and green lines, respectively.
See also Figure S2.