Figure 6.
Apigenin and Luteolin Induce Intron Retention with Weak Splice Sites
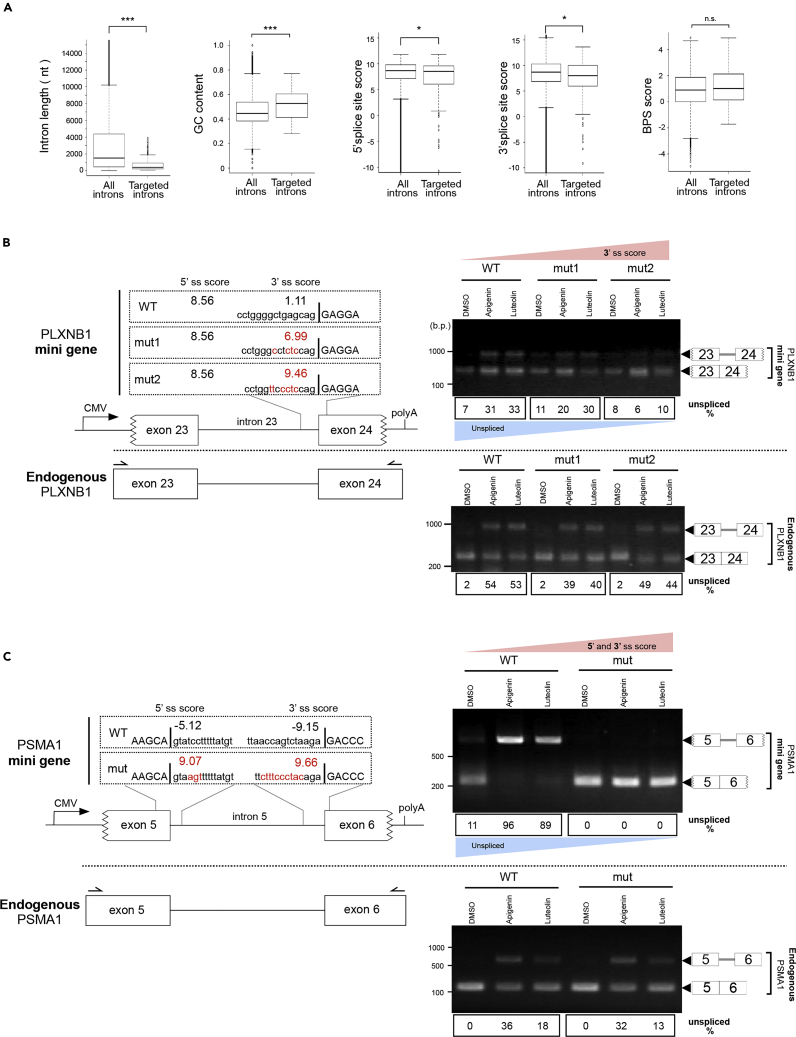
(A) The bioinformatic analysis on retained introns induced by apigenin and luteolin. All introns of human reference genes and retained introns induced by apigenin and luteolin were analyzed. Boxes show median (center line) and upper and lower quartile ranges. Whiskers show the lowest and highest values. Statistical analysis was performed using the two-sided Mann–Whitney U test. *p < 0.05, ***p < 0.001, n.s.: not significant.
(B and C) Upper left panels in (B and C) present schematic representations of mini gene constructs affected by apigenin and luteolin. The CMV promoter and BGH poly(A) sites are shown. Jagged line in the figure of the exon indicates that the edges of the exons have been partially deleted in order to distinguish endogenous mRNA from transgene-derived mRNA. The mutated nucleotides in each mutated-mini gene construct are shown in red, and the exon sequence is shown in upper case. Splice site score (ss score) was calculated using MaxEntScan. Lower left panels in (B and C) present schematic representations of the endogenous gene. The locations of the primers that amplify the endogenous target are marked in the schematic representation of the endogenous gene (black arrows). Right panels in (B and C) show the results of RT-PCR. DNA size in base pairs (b.p.) is indicated on the left side. The mRNAs derived from the transgene are shown in the upper right panels, and endogenous mRNAs are shown in the lower right panels. The digit panels below the photo show the percentage of unspliced mRNA band intensity and representative results of triplicate experiments.
See also Figure S8.