Abstract
Emerging evidence suggests that oroxylin A exhibits antitumor effects by inducing cell apoptosis. However, the involved molecular mechanisms have not been elucidated. Here we report that the apoptosis induced by oroxylin A was dependent on p62-mediated activation of caspase-8 in hepatocellular carcinoma cells. Furthermore, oroxylin A also caused p62/SQSTM1 proteolysis at Asp329 by activating caspase-8. Further studies confirm that mutation in p62 (D329H and D329G) was resistant to oroxylin A-mediated p62 cleavage and apoptosis. Due to the absence of the KIR domain that interacts with Keap1, the cleaved p62 reduced the stability of Nrf2, thereby causing oxidative stress and increasing ROS levels. In vivo, p62 similarly contributed to oroxylin A-exerted antitumor effect in xenograft model inoculated SMMC-7721 tumor. In conclusion, our findings indicated that oroxylin A triggered apoptosis through caspase-8 activation and p62/SQSTM1 proteolysis.
Keywords: Oroxylin A, Apoptosis, Caspase-8, p62/SQSTM1, Proteolysis
Abbreviations: SQSTM1, Sequestosome-1; UBA, ubiquitin-associated; LIR, LC3-interacting region; aPKCs, atypical protein kinases C; RIP, receptor interacting protein; TRAF6, tumor necrosis factor receptor-associated factor 6; RAG, Ras-related guanosine triphosphate (GTP) binding; mTORC1, mechanistic target of rapamycin complex 1; PB1, Phox-Bem1p; DISC, death-inducing signaling complex; OA, oroxylin A
1. Introduction
The scaffold protein p62, also known as SQSTM1, is involved in the autophagy degradation of ubiquitinated proteins [1,2]. p62 serves as a multifunctional molecule that regulates the key signalling pathways in cancer [3]. On the one hand, p62 protects cancer cells from cell death caused by different types of stimuli, including oxidative stress, so as to maintain the progress of cancer. On the other hand, p62 is positively correlated with cell death [[4], [5], [6], [7]]. Therefore, p62 has multifaceted functions and plays a complex role in determining the outcome of tumorigenesis.
The multifaceted functions of p62 depends on its different domains, which promote its interaction with different proteins. For example, p62 binds to ubiquitinated proteins through its ubiquitin-associated (UBA) domain and is anchored to LC3 in autophagosome membranes via its LC3-interacting region (LIR) domain [8]. In addition, p62 promotes oxidative stress response through the E3 ligase kelchlike ECH-associated protein 1 (Keap1), regulates NF-κB activation through atypical protein kinases C (aPKCs), receptor interacting protein (RIP) and tumor necrosis factor (TNF) receptor-associated factor 6 (TRAF6), regulates cell growth and anabolism through the Ras-related guanosine triphosphate (GTP) binding (RAG) family of guanosine triphosphatases (GTPases) that regulate the mechanistic target of rapamycin complex 1 (mTORC1) [[9], [10], [11]]. The N terminal of p62 protein also includes a Phox-Bem1p (PB1) domain, which mediates homo-oligomerization and other protein-protein interactions. In addition to these known functions, more and more studies believe that p62 is an important molecule linking autophagy and apoptosis. Studies have found that p62-mediated caspase 8-dependent apoptosis, but its molecular mechanism is still unclear.
Caspase-8 is a key protein in the extrinsic apoptotic pathway and its activation depends on oligomerization and self-cleavage. After activation, full-length caspase-8 is cleaved into p43/41 and p10 fragments, which are released into the cytoplasm to activate downstream caspases. Typical caspase-8 activation pathways rely on cell-surface death receptors through the death-inducing signaling complex (DISC) [12]. Therefore, recent studies have shown that TRAIL can induce p62-dependent activation of caspase-8, and the accumulation of p62 can also promote the activation of caspase-8 [13]. In addition, the activation of caspase-8 has been confirmed to cleave p62 protein and ultimately activate mTOR [14]. Therefore, the relationship between p62 and caspase-8 is very difficult to understand, and the exploration of the regulatory mechanism of p62 and caspase-8 helps to clarify the correlation between autophagy and apoptosis.
Oroxylin A (C16H12O5) is a flavonoid compound isolated from the root of scutellaria baicalensis, a traditional Chinese herbal medicine. It has been reported that Oroxylin A has antioxidant, anti-inflammatory and anti-tumor activities [[15], [16], [17]]. In terms of anti-tumor effect, Oroxylin A has been proved to have the effect of inducing autophagy and apoptosis [18]. But the molecular mechanisms involved remain unclear. Our study found that the apoptosis induction effect of Oroxylin A is dependent on the p62-mediated activation of caspase-8.
2. Results
2.1. Oroxylin A inhibits cell growth in human cancer cells
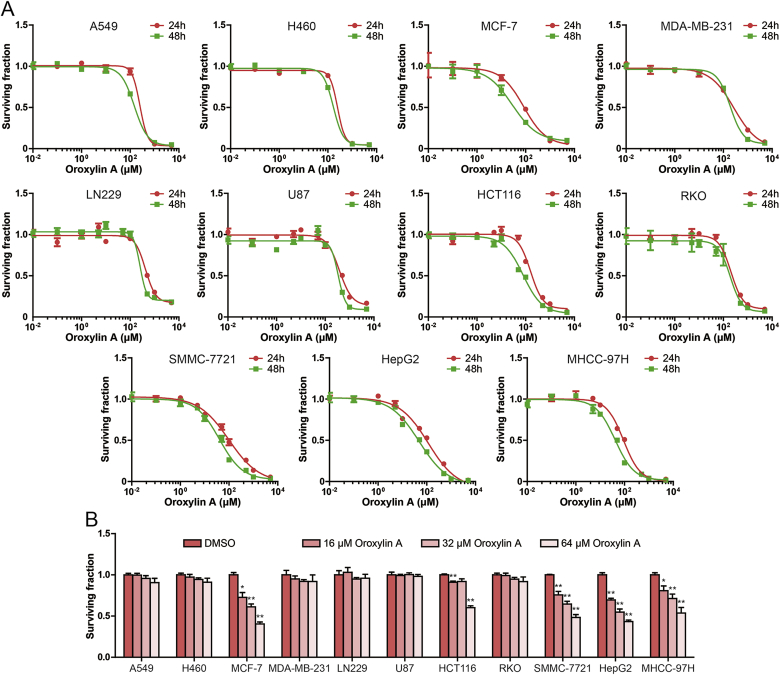
To evaluate the antitumor activity of oroxylin A. The inhibitory effects of oroxylin A on the growth of human non-small cell lung cancer cells A549 and H460, breast cancer cells MCF-7 and MDA-MB-231, glioma cells LN229 and U87, colon cancer cells HCT116 and RKO and hepatocellular carcinoma cells SMMC-7721, HepG2 and MHCC-97H were studied by MTT assay. As shown in Fig. 1A, hepatocellular carcinoma cells are more sensitive to oroxylin A than other types of tumor cells, excluding MCF-7 cells and HCT116 cells. In addition, we found that the growth inhibition effect of oroxylin A for 24 h treatment on tumor cells was not as effective as 48 h treatment. The IC50 values of oroxylin A in SMMC-7721, HepG2 and MHCC-97H cells were 40.34, 48.58 and 41.02 μM for 48 h treatment, respectively. Then, we examined the effect of 16, 32 and 64 μM oroxylin A on cell survival. As shown in Fig. 1B, oroxylin A significantly inhibited hepatocellular carcinoma cell survival, but had no significant effect on other cell survival, excluding MCF-7 cells and HCT116 cells.
Fig. 1.
Oroxylin A suppressed human cancer cell growth. (A) MTT assay was used to detect cell surviving fraction after treatment of different concentrations of oroxylin A for 24 h and 48 h in A549, H460, MCF-7, MDA-MB-231, LN229, U87, HCT116, RKO, SMMC-7721, HepG2 and MHCC-97H cells. (B) Effect of certain concentration oroxylin A on the surviving fraction in human cancer cell were measured by MTT assay. Data are presented as mean ± SD. *P < 0.05, **P < 0.01 compared with DMSO group.
2.2. Oroxylin A induces apoptosis in hepatocellular carcinoma cells
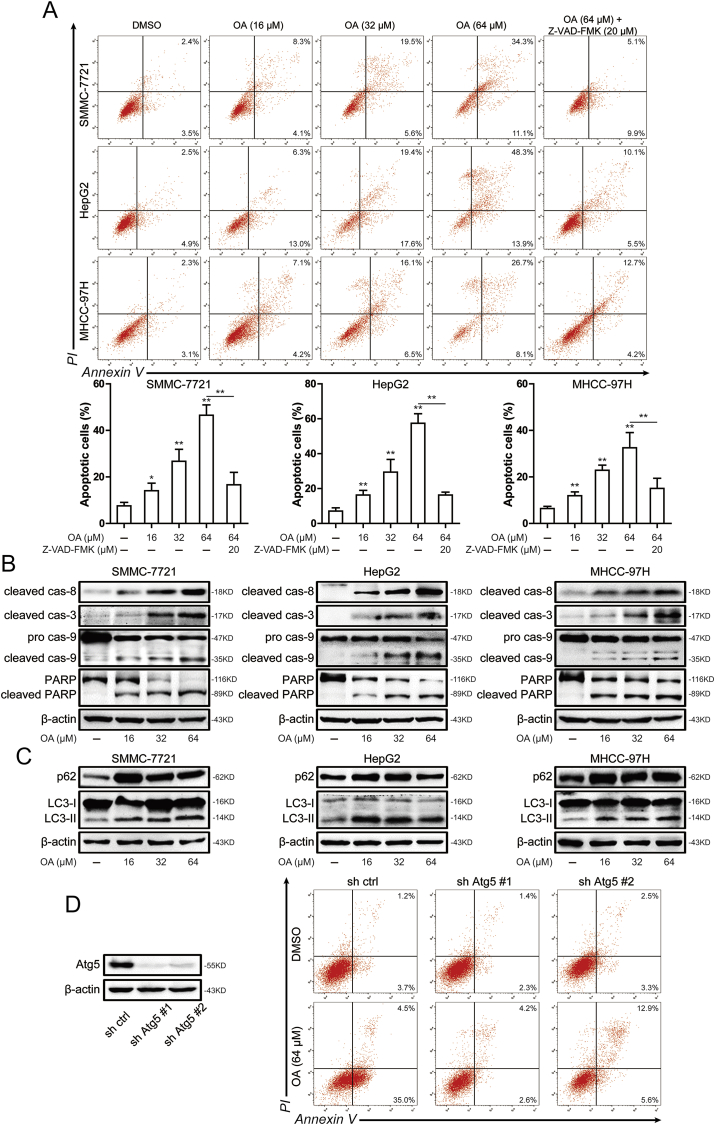
To detect the apoptosis induced by oroxylin A in hepatocellular carcinoma cells, Annexin V/PI staining assay was used. As shown in Fig. 2A, increased apoptotic was detected after oroxylin A treatment for 48 h. The percentage of apoptotic cells were 46.5% ± 3.6%, 57.4% ± 4.5%, and 32.5% ± 5.4% after treated with 64 μM oroxylin A in SMMC-7721, HepG2 and MHCC-97H cells, respectively. After the treatment of pan-caspase inhibitor Z-VAD-FMK, apoptotic cells induced by oroxylin A were reduced. Furthermore, the apoptosis related proteins such as caspase-8, caspase-9, caspase-3 and PARP were investigated by western blots. After treatment with oroxylin A for 48 h, the cleaved caspase-8, cleaved caspase-9, cleaved caspase-3 and cleaved PARP expression increased in a concentration-dependent manner (Fig. 2B). These results showed that oroxylin A induced apoptosis in hepatocellular carcinoma cells. Studies have shown that oroxylin A induced autophagy in the upstream of apoptosis and promotes apoptosis of hepatocellular carcinoma cells [18]. Therefore, we investigated autophagy induced by oroxylin A. As shown in Fig. 2C, the expression of p62 and LC3-II were up-regulated after 16 μM oroxylin A treatment. However, the expression of p62 and LC3-II were not further increased with the increase of oroxylin A concentration. To confirm the effect of autophagy on oroxylin A-induced apoptosis, we tested apoptosis induced by oroxylin A after knockdown of ATG5, which was considered to crucial function around later events of autophagosome formation [19]. After knockdown of ATG5, oroxylin A-induced apoptosis was decreased in hepatocellular carcinoma cells (Fig. 2D). Therefore, oroxylin A-mediated apoptosis was blocked by ATG5 knockdown in hepatocellular carcinoma cells.
Fig. 2.
Oroxylin A induced apoptosis in hepatocellular carcinoma cells. (A) Induction of apoptosis was measured by Annexin-V/PI double-staining assay in SMMC-7721, HepG2 and MHCC-97H cells after 16, 32 and 64 μM oroxylin A or 20 μM Z-VAD-FMK treatment (top). Histograms shows the distribution of apoptotic cells for three independent experiments (mean ± SD) (bottom). (B) The levels of cleaved caspase-8, cleaved caspase-3, caspase-9 and PARP were assessed by western blot in SMMC-7721, HepG2 and MHCC-97H cells after 16, 32 and 64 μM oroxylin A treatment. (C) The levels of p62 and LC3 were assessed by western blot in SMMC-7721, HepG2 and MHCC-97H cells after 16, 32 and 64 μM oroxylin A treatment. (D) The level of ATG5 was assessed by western blot in SMMC-7721 cells after knockdown of ATG5 (left). Induction of apoptosis was measured by Annexin-V/PI double-staining assay in SMMC-7721 cells after knockdown of ATG5 (right). Data are presented as mean ± SD. *P < 0.05, **P < 0.01 compared with DMSO group.
2.3. Caspase-8 activation contributes to oroxylin A-induced apoptosis
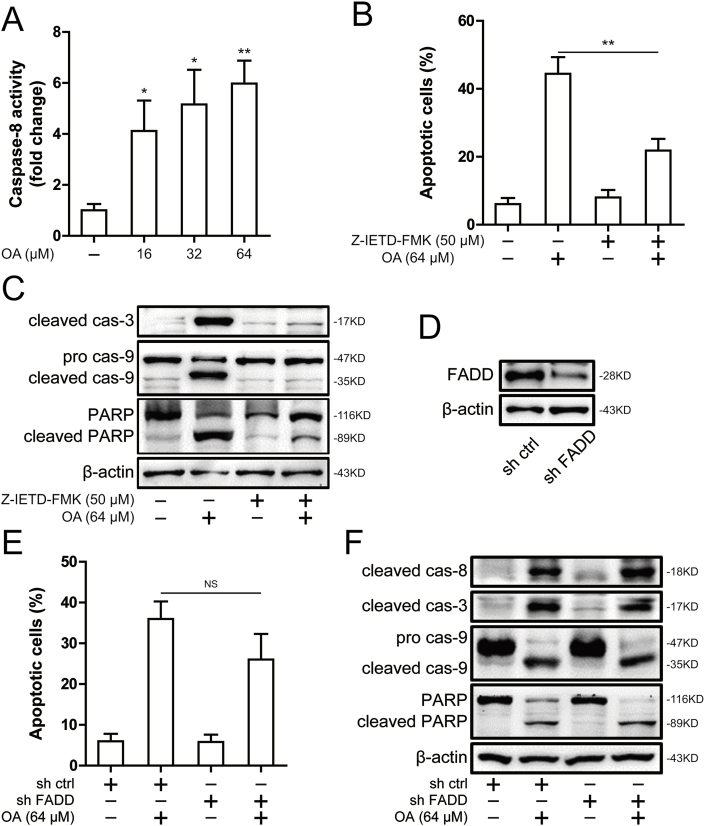
Oroxylin A was shown to cleave caspase-8 (Fig. 2B) and was also found to significantly activate caspase-8 using a caspase-8 activity assay (Fig. 3A). To confirm the critical role of caspase-8 as a mediator of oroxylin A-induced apoptosis, we used Z-IETD-FMK, a caspase-8 inhibitor, where oroxylin A-induced apoptosis was blocked (Fig. 3B). Furthermore, caspase-3, caspase-9 and PARP cleavage was attenuated after treatment of Z-IETD-FMK (Fig. 3C). This suggests that caspase-8 activation was involved in oroxylin A-induced apoptosis. Caspase-8 has been shown to be activated by self-aggregation in response to death receptor stimulation upon ligand binding that enables its recruitment to the DISC [20]. To exclude the possibility that caspase-8 activation is mediated by the DISC, we utilized FADD shRNA for FADD knockdown (Fig. 3D). Accordingly, oroxylin A-induced apoptosis and cleavage of caspase-8, caspase-3, caspase-9 and PARP was also not affected by FADD knockdown (Fig. 3E and F). In summary, the activation of caspase-8, which is independent of FADD, contributed to oroxylin A-induced apoptosis.
Fig. 3.
Caspase-8 mediated the apoptosis induced by oroxylin A in SMMC-7721 cells. (A) The caspase-8 activity was measured in SMMC-7721 cells after 16, 32 and 64 μM oroxylin A treatment. (B) Induction of apoptosis was measured by Annexin-V/PI double-staining assay in SMMC-7721 cells after treatment of 50 μM Z-IETD-FMK and 64 μM oroxylin A. (C) The levels of cleaved caspase-3, caspase-9 and PARP were assessed by Western blot in SMMC-7721 cells after treatment of 50 μM Z-IETD-FMK and 64 μM oroxylin A. (D) The levels of FADD was assessed by Western blot in SMMC-7721 cells after knockdown of FADD. (E) Induction of apoptosis was measured by Annexin-V/PI double-staining assay in SMMC-7721 cells after knockdown of FADD. (F) The levels of cleaved caspase-8, cleaved caspase-3, caspase-9 and PARP were assessed by Western blot in SMMC-7721 cells after knockdown of FADD. Data are presented as mean ± SD. *P < 0.05, **P < 0.01 compared with DMSO group.
2.4. p62 is involved in oroxylin A-induced apoptosis and caspase-8 activation in hepatocellular carcinoma cells
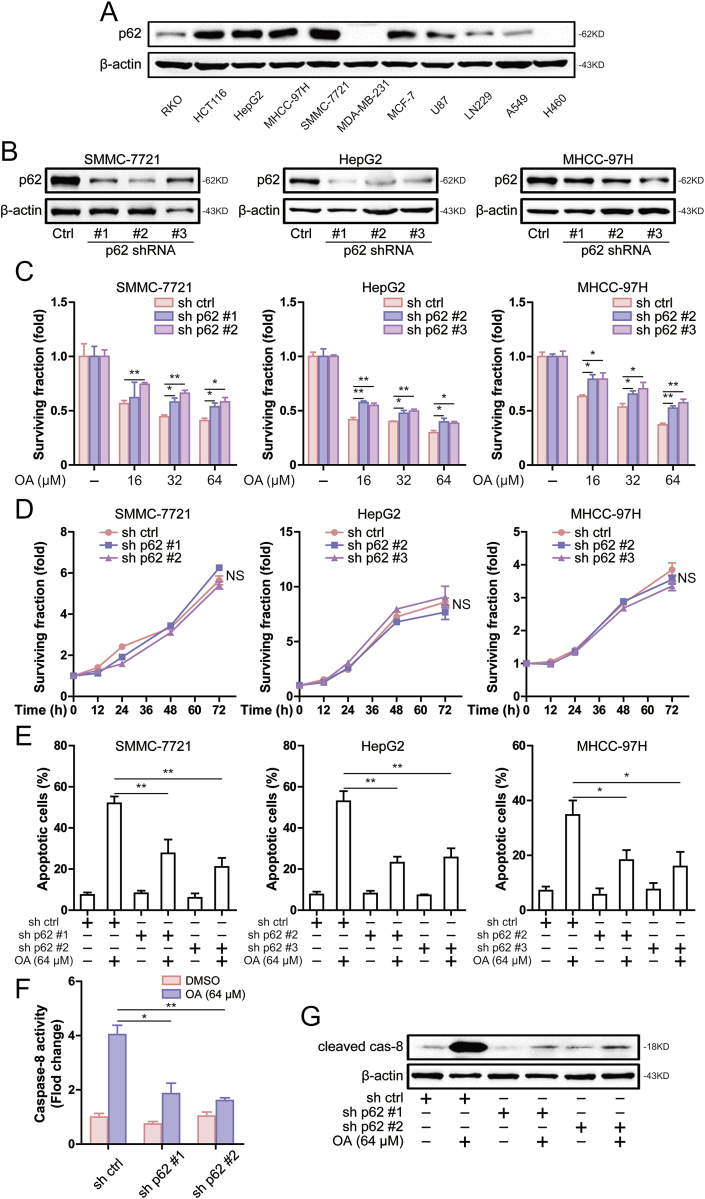
In addition to the DISC-mediated caspase-8 activation, p62 has also been reported to activate caspase-8 [6,21]. Basic expression levels of p62 protein in different cell lines were detected and showed that p62 was highly expressed in HCT116, SMMC-7721, HepG2, MHCC-97H and MCF-7 cells (Fig. 4A). By comparing the results in Fig. 1, it is not difficult to find that p62 is highly expressed in cells sensitive to oroxylin A. Therefore, we speculated that p62 mediated apoptosis induced by oroxylin A. To clarify the role of p62 in oroxylin A-induced apoptosis, we reduced the expression of p62 in hepatocellular carcinoma cells (Fig. 4B). Knockdown of p62 resulted in decreased sensitivity to oroxylin A in hepatocellular carcinoma cells (Fig. 4C). In addition, knockdown of p62 did not affect cell growth (Fig. 4D). This suggests that the decreased sensitivity to oroxylin A induced by knockdown of p62 was not caused by the difference in cell growth. Furthermore, oroxylin A-induced apoptosis was attenuated after knockdown of p62 (Fig. 4E). In order to explore the mechanism of p62 in oroxylin A-induced apoptosis, we examined whether p62 interferes with caspase-8 activation. As shown in Fig. 4F, knockdown of p62 decreased caspase-8 activity induced by oroxylin A. Furthermore, caspase-8 cleavage was attenuated after knockdown of p62 (Fig. 4G). These results suggested that p62 was involved in oroxylin A-induced apoptosis and caspase-8 activation in hepatocellular carcinoma cells.
Fig. 4.
Knockdown of p62 attenuates oroxylin A-induced apoptosis and caspase-8 activation in hepatocellular carcinoma cells. (A) The levels of p62 was assessed by western blot in A549, H460, MCF-7, MDA-MB-231, LN229, U87, HCT116, RKO, SMMC-7721, HepG2 and MHCC-97H cells. (B) The levels of p62 was assessed by western blot in SMMC-7721, HepG2 and MHCC-97H cells after transfection of p62 shRNA. (C) Effect of certain concentration oroxylin A on the surviving fraction were measured by MTT assay in SMMC-7721, HepG2 and MHCC-97H cells after knockdown of p62. (D) MTT assay was used to detect cell surviving fraction after culture for 12 h, 24 h, 48 h and 72 h in SMMC-7721, HepG2 and MHCC-97H cells. (E) Induction of apoptosis was measured by Annexin-V/PI double-staining assay in SMMC-7721, HepG2 and MHCC-97H cells after knockdown of p62. (F) The caspase-8 activity was measured in SMMC-7721 cells after knockdown of p62. (G) The levels of cleaved caspase-8 was assessed by western blot in SMMC-7721 cells after knockdown of p62. Data are presented as mean ± SD. *P < 0.05, **P < 0.01.
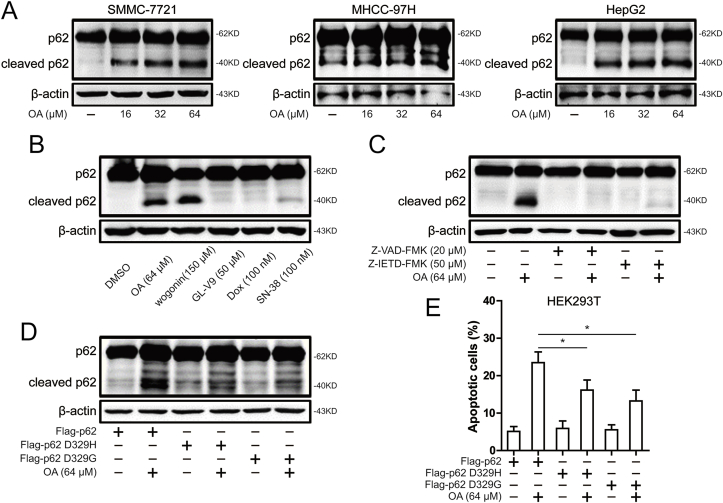
2.5. Caspase-8 activation induced by oroxylin A causes the cleavage of p62
The apoptosis induced by oroxylin A was further increased with the increase of oroxylin A concentration in hepatocellular carcinoma cells (Fig. 2A). While oroxylin A was shown to increase p62 expression at 16 μM, but does not further increase the expression of p62 with the increase of oroxylin A concentration (Fig. 2C). There seems to be a discrepancy, but studies have found proteolytic cleavage of p62 by caspase-8 determines whether p62 functions as an mTORC1 signaling adaptor or autophagy receptor [22]. Therefore, the cleavage of p62 was detected after oroxylin A treatment. As shown in Fig. 5A, the appearance of a shorter (~40 kDa) p62 protein on oroxylin A concentration for long exposure. Compared with other flavonoids and chemotherapy drugs, the cleavage of p62 depends on the treatment of oroxylin A or wogonin (Fig. 5B). Moreover, the cleavage of p62 was diminished after treatment of Z-VAD-FMK or Z-IETD-FMK (Fig. 5C). Studies have found that two mutations in p62 (D329H and D329G) that affect the caspase-8 cleavage site and therefore abolish p62 cleavage generation. Therefore, immunoblots from HEK293T cells transfected with Flag-p62 WT, Flag-p62 D329H or D329G variant and left treated with oroxylin A. As shown in Fig. 5D, p62 D329H and D329G variant were resistant to oroxylin A–mediated cleavage. Furthermore, oroxylin A-induced apoptosis was attenuated in HEK293T cells after transfection of Flag-p62 D329H or D329G variant (Fig. 5E). These results indicated that caspase-8–mediated cleavage of p62 was involved in oroxylin A-induced apoptosis.
Fig. 5.
Oroxylin A induced caspase-8 to cleave p62. (A) The levels of p62 and cleaved p62 were assessed by western blot in SMMC-7721, HepG2 and MHCC-97H cells after 16, 32 and 64 μM oroxylin A treatment. (B) The levels of p62 and cleaved p62 were assessed by western blot in SMMC-7721 cells after 64 μM oroxylin A, 150 μM wogonin, 50 μM GL-V9, 100 nM doxorubicin and 100 nM SN-38 treatment. (C) The levels of p62 and cleaved p62 were assessed by western blot in SMMC-7721 cells after 64 μM oroxylin A treatment or combined with 20 μM Z-VAD-FMK or 50 μM Z-IETD-FMK. (D) The levels of p62 were assessed by western blot in HEK293T cells after transfection with Flag-p62 WT, Flag-p62 D329G or D329H variant. (E) Induction of apoptosis was measured by Annexin-V/PI double-staining assay in HEK293T cells after transfection with Flag-p62 WT, Flag-p62 D329G or D329H variant. Data are presented as mean ± SD. *P < 0.05, **P < 0.01.
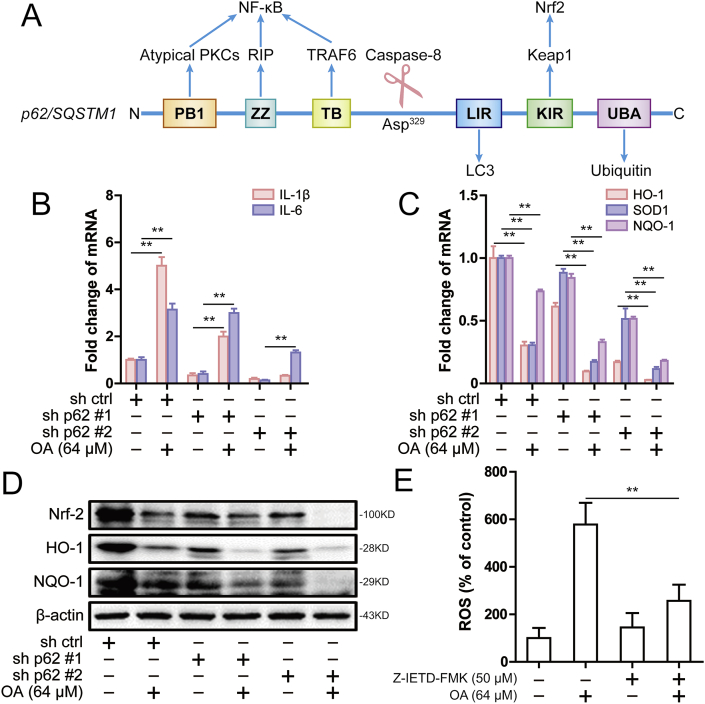
2.6. Oroxylin A-triggered p62 cleavage induces oxidative stress through down-regulation of Nrf2
Extensive studies have confirmed that p62 promotes NF-κB activation through atypical protein kinases C (aPKCs), receptor interacting protein (RIP) and tumor necrosis factor (TNF) receptor-associated factor 6 (TRAF6), and promotes Nrf2 activation through the E3 ligase kelchlike ECH-associated protein 1 (KEAP1) (Fig. 6A) [9,23]. Therefore, we next studied the effect of oroxylin A on NF-κB and Nrf2 signaling pathway. By detecting the transcription downstream genes of NF-κB and Nrf2 signaling pathway, it was found that oroxylin A could up-regulate the mRNA levels of IL-1β and IL-6, and down-regulate the mRNA levels of HO-1, SOD1 and NQO-1 (Fig. 6B and C). Similarly, oroxylin A significantly down-regulated Nrf2, HO-1 and NQO1 expression (Fig. 6D). These results suggested that oroxylin A down-regulated Nrf2 signaling pathway. In previous studies, the caspase-8 cleavage site was narrowed down to Asp329, which was between the TB domain and the LIR domain (Fig. 6A) [11]. Therefore, the reason for this result may be that oroxylin A-triggered p62 cleavage loses its binding site with Keap1 and thus inhibits Nrf2 signaling pathway. The inhibition of Nrf2 signaling pathway leads to increased oxidative stress. Therefore, it was found that oroxylin A increased the ROS level in SMMC-7721 cells, while inhibition of caspase-8 activation reversed oroxylin A-mediated the increase of ROS level (Fig. 6E). In conclusion, oroxylin A induced oxidative stress and down-regulated Nrf2 by triggering p62 cleavage.
Fig. 6.
Oroxylin A-driven p62 cleavage down-regulated Nrf2 to induce oxidative stress. (A) Domain composition of human p62/SQSTM1 showing the caspase-8 cleavage site (not to scale). Blue arrows point to regions for interactions with the indicated proteins. (B) The IL-1β and IL-6 mRNA was measured by real-time PCR following 64 μM oroxylin A treatment or p62 knockdown in SMMC-7721 cells. (C) The HO-1, SOD1 and NQO-1 mRNA was measured by real-time PCR following 64 μM oroxylin A treatment or p62 knockdown in SMMC-7721 cells. β-actin was used as internal control. The relative levels were calculated to β-actin mRNA expression. (D) The levels of Nrf2, HO-1 and NQO-1 were assessed by western blot following 64 μM oroxylin A treatment or p62 knockdown in SMMC-7721 cells. (E) The ROS level was assessed following 64 μM oroxylin A and 50 μM Z-IETD-FMK treatment in SMMC-7721 cells. Data are presented as mean ± SD. *P < 0.05, **P < 0.01. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
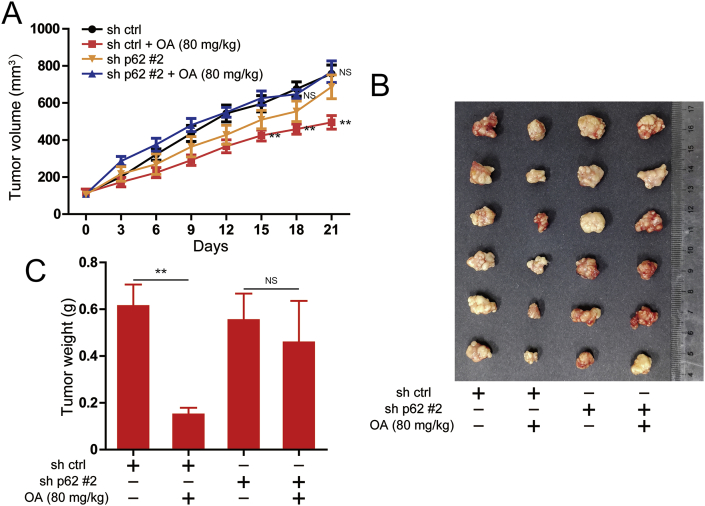
2.7. p62 is involved in oroxylin A-exerted antitumor effect in xenograft mice inoculated SMMC-7721 tumor
A xenograft model transplanted with SMMC-7721 tumor was applied to evaluate the antitumor effect of oroxylin A in vivo. At the same measurement day, the tumor volume of oroxylin A treatment group mice was smaller than control group mice, while after knockdown of p62, the tumor volume of oroxylin A treatment group mice was not significantly different from that of control group mice (Fig. 7A). The tumor size was also visually smaller in oroxylin A treatment group, but tumor size did not appear to change significantly after knockdown of p62 (Fig. 7B). Moreover, the tumor weight of oroxylin A-treated mice was significantly smaller than control group, while the tumor weight did not change after knockdown of p62 (Fig. 7C). Taken together, all these results suggested that p62 mediates the inhibitory effect of oroxylin A on tumor growth.
Fig. 7.
Effects of oroxylin A on the tumorigenicity in SMMC-7721 cells in vivo. (A) Tumor volume of control and oroxylin A treatment groups in SMMC-7721 cells or in knockdown of p62 SMMC-7721 cells were measured and calculated once every 3 days. (B) Images of SMMC-7721 tumors from indicated group on day 21. (C) Weight of tumor in control and oroxylin A treatment groups. Data are presented as mean ± SD. *P < 0.05, **P < 0.01.
3. Discussion
Oroxylin A has been proved to have a significant anti-tumor effect, but the anti-tumor mechanism is still under further study [[24], [25], [26], [27]]. In order to evaluate the antitumor activity of oroxylin A, we tested the inhibitory effect of oroxylin A on cell growth in different tumor cell lines. Results showed that oroxylin A significantly inhibited the cell growth in hepatocellular carcinoma cells. Further studies showed that oroxylin A promoted caspase-8-mediated apoptosis. P62 was involved in caspase-8 aggregation to mediate extrinsic apoptosis signaling [28]. Therefore, we examined the expression of p62 in different tumor cell lines. The results showed that cells with high expression of p62 were more sensitive to oroxylin A than cells with low expression of p62. After knockdown of p62 expression, the apoptosis and caspase-8 activation induced by oroxylin A were significantly inhibited. In addition, we also found that oroxylin A caused the cleavage of p62, which was dependent on the proteolytic effect of caspase-8. Due to the absence of the KIR domain that interacts with Keap1, the cleaved p62 reduced the stability of Nrf2 and further induces oxidative stress [29,30]. Together these data indicated that oroxylin A triggered apoptosis through caspase-8 activation and p62/SQSTM1 proteolysis in hepatocellular carcinoma cells.
In our study we examined the effects of oroxylin A on cell growth in different tumor cell lines, we found that hepatocellular carcinoma cell lines such as SMMC-7721, HepG2 and MHCC-97H, breast cancer cell line MCF-7 and colorectal cancer cell line HCT116 were sensitive to oroxylin A (Fig. 1). Subsequent studies found that the sensitivity of oroxylin A may be related to the level of p62 expression. Therefore, we decreased the p62 expression in hepatocellular carcinoma cell lines and found that the sensitivity of oroxylin A was reduced after knockdown the p62 expression (Fig. 4C and E). Further studies confirmed that knockdown of p62 suppressed caspase-8 activation induced by oroxylin A (Fig. 4F and G). The above results suggest that p62 may be the key molecule to determine the anti-tumor effect of oroxylin A.
Autophagy is thought to be an intracellular degradation system that delivers cytoplasmic proteins to lysosomes [31]. In our study, oroxylin A-induced apoptosis was inhibited after ATG5 knockdown (Fig. 2D). This result indicated that autophagy was related to oroxylin A-induced apoptosis, which was consistent with the results of 3-MA inhibiting apoptosis induced by oroxylin A [18]. While p62 is itself degraded by autophagy and may serve to link ubiquitinated proteins to the autophagy degradation in the lysosome. Since p62 accumulates when autophagy is inhibited, and decreased levels can be observed when autophagy is induced [32]. Therefore, ATG5 knockdown-mediated p62 accumulation and subsequent Nrf2 activation leads to the decrease of oroxylin A-induced apoptosis.
The involvement of p62 in caspase-8 activation has been confirmed by more and more studies. Meanwhile, p62 has also been shown to be cleaved by caspase-6 and caspase-8 in vitro [33]. In our study, it is accidentally found that oroxylin A caused the cleavage of p62 by proteolytic cleavage of caspase-8 (Fig. 5A). In addition to oroxylin A, wogonin, a flavonoid compound isolated from scutellaria baicalensis root, also caused the cleavage of p62. Interestingly, GL-V9, a derivative of wogonin, and the chemotherapy drugs doxorubicin and SN-38 failed to cause the cleavage of p62 (Fig. 5B). Additionally, p62 can promote the activation of NF-κB and Nrf2 [34]. However, oroxylin A promoted the transcription of NF-κB downstream target genes and inhibited the transcription of Nrf2 downstream target genes (Fig. 6B and C). These results may be due to the fact that the cleavage of p62 by caspase-8 maintained only the domains associated with NF-κB activation, such as PB1, ZZ, and TB, while KIR, a domain interacted with Keap1, was cleaved away that lose the function to stabilize Nrf2. The instability of Nrf2 triggered by oroxylin A caused oxidative stress, which increased ROS levels. This may be the reason why oroxylin A promoted caspase-9 activation.
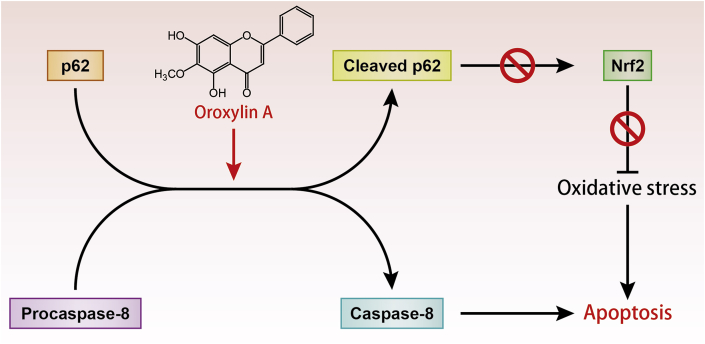
In conclusion, the present study demonstrated that oroxylin A triggered apoptosis through caspase-8 activation and p62/SQSTM1 proteolysis in hepatocellular carcinoma cells. We provide evidence that p62 was the key protein of oroxylin A-induced apoptosis. On the one hand, p62 promoted caspase-8 activation. On the other hand, caspase-8 caused p62 cleavage, which leads to oxidative stress. This implies that oroxylin A may be a potential treatment for hepatocellular carcinoma (Fig. 8). However, the integrated safety assessment and optimization of treatment options in clinical application of oroxylin A deserve further study.
Fig. 8.
The possible mechanism of oroxylin A inducing autophagy-related apoptosis. Oroxylin A promotes caspase-8 activation and p62 proteolysis through the interaction of procaspase-8 and p62. On the one hand, caspase-8 activation triggered apoptosis. On the other hand, cleaved p62 inhibited Nrf2 to generate oxidative stress and eventually triggered apoptosis.
4. Materials and methods
4.1. Reagents and antibodies
Oroxylin A was isolated from the root of Scutellaria baicalensis as previously described [35]. Samples containing oroxylin A at a minimum of 99% purity were used for the experiments unless otherwise indicated. Oroxylin A was dissolved in dimethylsulfoxide (DMSO) as a stock solution, stored at −20 °C, and freshly diluted with medium to the final concentration in vitro study.
DMSO was purchased from Sigma-Aldrich (St. Louis, USA). BSA was purchased from Roche Diagnosis Ltd. (Shanghai, China).
Primary antibodies against to caspase-8, caspase-9, cleaved-caspase-3, PARP, FADD, NQO-1 and β-actin were obtained from ABclonal (ABclonal, Wuhan, China). Antibodies against to Nrf-2 was from Bioworld (Bioworld, OH, USA) and antibodies against to LC3 and HO-1 were purchased from Cell Signaling Technology (CST, MA, USA). Antibodies against to SQSTM1/p62 was from Abcam (Abcam, Cambridge, UK). HRP Goat Anti-Mouse IgG (H + L) and HRP Goat Anti-Rabbit IgG (H + L) were from ABclonal (ABclonal, Wuhan, China). High-sig ECL Western Blotting Substrate was from Tanon (Tanon, Shanghai, China).
4.2. Cell culture
Human non-small cell lung cancer A549 cells and H460 cells, human breast cancer MCF-7 cells and MDA-MB-231 cells, human glioma LN229 cells and U87 cells, human colon cancer HCT116 cells and RKO cells, human hepatocellular carcinoma SMMC-7721 cells, HepG2 cells and MHCC-97H cells, HEK293T cells were obtained from Cell Bank, Chinese Academy of Sciences. A549 cells were cultured in F-12 medium (Gibco, Waltham, USA). H460, SMMC-7721 and MHCC-97H cells were cultured in RPMI-1640 medium (Gibco, Carlsbad, USA). MCF-7, MDA-MB-231, LN229, U87, HCT116, RKO, HepG2 and HEK293T cells were cultured in DMEM medium (Gibco, Carlsbad, USA). Medium was supplemented with 10% (v/v) fetal bovine serum (Gibco, Carlsbad, USA) and 0.05 mM 2-mercaptoethanol, 100 U/ml benzyl penicillin and 100 mg/m streptomycin. Cells were cultured in a humidified environment with 5% CO2 at 37 °C.
4.3. MTT assay
Experiments were done in triplicate in a parallel manner for each concentration of Oroxylin A used and the results are presented as mean ± SEM. Control cells were given culture media containing 0.1% DMSO. After incubation for 24 or 48 h, 20 μL of 5 mg/mL MTT was added to cells, and cells were incubated at 37 °C for another 4 h. The absorbance (A) was measured at 570 nm using an ELx800 automated microplate reader (BioTek Instruments, Inc.). The surviving fraction was calculated using the following equation: surviving fraction = average absorbance of treated group/average absorbance of control group × 100%. IC50 values were taken as the concentration that caused 50% surviving fraction and was calculated by the Logit method.
4.4. Annexin V/PI staining
Cells were harvested, washed, and resuspended in PBS, then stained with the Annexin V/PI Cell Apoptosis Detection Kit (KeyGen Biotech, Nanjing, China) according to the manufacturer's instructions. Data acquisition and analysis were performed with a Becton Dickinson FACS Calibur flow cytometer using Cell-Quest software (BD Biosciences, Franklin Lakes, USA). The cells in early stages of apoptosis were Annexin V positive and PI negative, whereas the cells in the late stages of apoptosis were both Annexin V and PI positive.
4.5. Cell transfection and lentivirus package
The shRNA targeting human ATG5, p62 or control shRNA with scrambled sequence, pMD2G and psPAX2 plasmids were transfected into HEK293T cells using Lipofectamine 2000™ reagent (Invitrogen, CA, USA), according to the manufacturer's instructions. Virus supernatant was collected 48 h after transfection, and then infected target cells with virus supernatant and selected puromycin for screening. Sequences of specific shRNAs (from Sigma shRNA Mission library) used in this study are as follows:
sh p62#1 (5’-CCGGCCTCTGGGCATTGAAGTTGATCTCGAGATCAACTTCAATGCCCAGAGGTTTTT-3’);
sh p62#2 (5’-CCGGGCAGATGAGAAAGATCGCCTTCTCGAGAAGGCGATCTTTCTCATCTGCTTTTT-3’);
sh p62#3 (5’-CCGGCCGAATCTACATTAAAGAGAACTCGAGTTCTCTTTAATGTAGATTCGGTTTTT-3’);
sh Atg5#1 (5’-CCGGCCTGAACAGAATCATCCTTAACTCGAGTTAAGGATGATTCTGTTCAGGTTTTT-3’);
sh Atg5#2 (5’-CCGGAGATTGAAGGATCAACTATTTCTCGAGAAATAGTTGATCCTTCAATCTTTTTT-3’);
sh FADD (5’-CCGGGTGCAGCATTTAACGTCATATCTCGAGATATGACGTTAAATGCTGCACTTTTT-3’)
4.6. Western blot analysis
Western blot analysis was performed as previously described [36]. Protein samples were separated by 10% SDS-PAGE and transferred onto nitrocellulose membranes. The membrane was blocked with 3% no fat milk in PBST at 37 °C for 1 h and incubated with the indicated antibodies overnight at 4 °C, and then with HRP Goat Anti-Mouse IgG (H + L) or HRP Goat Anti-Rabbit IgG (H + L) secondary antibody for 1 h at 37 °C. The samples were visualized with High-sig ECL Western Blotting Substrate (Tanon, Shanghai, China) and Fully Automatic Chemiluminescence Image Analysis System (Tanon, Shanghai, China).
4.7. RNA isolation and qPCR
RNA was isolated from cells and reverse transcribed, and qPCR was performed as previously described [37]. Primer sequences are as follows:
IL-1β F: 5’-ATGATGGCTTATTACAGTGGCAA-3’; R: 5’-GTCGGAGATTCGTAGCTGGA-3’;
IL-6 F: 5’- ACTCACCTCTTCAGAACGAATTG-3’; R: 5’- CCATCTTTGGAAGGTTCAGGTTG-3’;
HO-1 F: 5’- AAGACTGCGTTCCTGCTCAAC-3’; R: 5’- AAAGCCCTACAGCAACTGTCG-3’;
SOD1 F: 5’- GGTGGGCCAAAGGATGAAGAG-3’; R: 5’- CCACAAGCCAAACGACTTCC-3’;
NQO-1 F: 5’- GAAGAGCACTGATCGTACTGGC-3’; R: 5’- GGATACTGAAAGTTCGCAGGG-3’;
4.8. Measurement of ROS levels
The level of ROS was detected using fluorescent dye 2, 7-dichlorofluorescein-diacetate (DCFHDA) obtained from Beyotime (Shanghai, China). Cells were collected and incubated with DCFH-DA for 30 min at 37 °C in the dark. The fluorescence intensity was measured using flow cytometry.
4.9. Animal studies
Female BALB/c nude mice, 35–40 days old and with weight ranging from 18 to 22 g, were supplied by Shanghai Laboratory Animal Limited Company. The mice were maintained in a pathogen-free environment (23 ± 2 °C and 55 ± 5% humidity) on a 12 h light–12 h dark cycle with food and water supplied ad libitum throughout the experimental period. Mice were subcutaneously inoculated with SMMC-7721 cells (7 × 106). Tumor volume (TV) was calculated using the following formula: TV (mm3) = d2 × D/2, where d and D are the shortest and the longest diameters, respectively. Animal study and euthanasia was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Committee on the Ethics of Animal Experiments of the China Pharmaceutical University.
4.10. Statistical analysis
The data shown in the study were obtained in at least three independent experiments and all results represent the mean ± S.D. Differences between the groups were assessed by one-way ANOVA test. Details of each statistical analysis used are provided in the figure legends. Differences with P values < 0.05 were considered statistically significant.
Declaration of competing interst
None declared.
Acknowledgments
This work was supported by the National Natural Science Foundation of China, China (No. 81903626, 81903625, 81872899, 81830105, 81673461), the National Science & Technology Major Project, China (No. 2017ZX09301014, 2018ZX09711001-005-023, 2018ZX09711001-003-007), the Project Program of State Key Laboratory of Natural Medicines, China (No.SKLNMZZCX201823), Social Development Project of Jiangsu Provincial Science and Technology Department, China (BE2018711), Natural Science Foundation of Jiangsu Province, China (BK20180576, BK20190563), “Double First-Class” University project, China (CPU 2018GF11, CPU2018GF05).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2019.101392.
Contributor Information
Yuan Gao, Email: 1620184502@cpu.edu.cn.
Na Lu, Email: nalu@cpu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Katsuragi Y., Ichimura Y., Komatsu M. p62/SQSTM1 functions as a signaling hub and an autophagy adaptor. FEBS J. 2015;282:4672–4678. doi: 10.1111/febs.13540. [DOI] [PubMed] [Google Scholar]
- 2.Lamark T., Svenning S., Johansen T. Regulation of selective autophagy: the p62/SQSTM1 paradigm. Essays Biochem. 2017;61:609–624. doi: 10.1042/EBC20170035. [DOI] [PubMed] [Google Scholar]
- 3.Moscat J., Karin M., Diaz-Meco M.T. p62 in cancer: signaling adaptor beyond autophagy. Cell. 2016;167:606–609. doi: 10.1016/j.cell.2016.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun X., Ou Z., Chen R., Niu X., Chen D., Kang R., Tang D. Activation of the p62-Keap1-NRF2 pathway protects against ferroptosis in hepatocellular carcinoma cells. Hepatology. 2016;63:173–184. doi: 10.1002/hep.28251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Umemura A., He F., Taniguchi K., Nakagawa H., Yamachika S., Font-Burgada J., Zhong Z., Subramaniam S., Raghunandan S., Duran A., Linares J.F., Reina-Campos M., Umemura S., Valasek M.A., Seki E., Yamaguchi K., Koike K., Itoh Y., Diaz-Meco M.T., Moscat J., Karin M. p62, Upregulated during preneoplasia, induces hepatocellular carcinogenesis by maintaining survival of stressed HCC-initiating cells. Cancer Cell. 2016;29:935–948. doi: 10.1016/j.ccell.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan X.Y., Zhong X.R., Yu S.H., Zhang L.C., Liu Y.N., Zhang Y., Sun L.K., Su J. p62 aggregates mediated Caspase 8 activation is responsible for progression of ovarian cancer. J. Cell Mol. Med. 2019;23:4030–4042. doi: 10.1111/jcmm.14288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zeng R.X., Zhang Y.B., Fan Y., Wu G.L. p62/SQSTM1 is involved in caspase-8 associated cell death induced by proteasome inhibitor MG132 in U87MG cells. Cell Biol. Int. 2014;38:1221–1226. doi: 10.1002/cbin.10311. [DOI] [PubMed] [Google Scholar]
- 8.Leong I. p62 - a new metabolic tumour suppressor. Nat. Rev. Endocrinol. 2018;14:324. doi: 10.1038/s41574-018-0018-0. [DOI] [PubMed] [Google Scholar]
- 9.Ichimura Y., Waguri S., Sou Y.S., Kageyama S., Hasegawa J., Ishimura R., Saito T., Yang Y., Kouno T., Fukutomi T., Hoshii T., Hirao A., Takagi K., Mizushima T., Motohashi H., Lee M.S., Yoshimori T., Tanaka K., Yamamoto M., Komatsu M. Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Mol. Cell. 2013;51:618–631. doi: 10.1016/j.molcel.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 10.Moscat J., Diaz-Meco M.T. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell. 2009;137:1001–1004. doi: 10.1016/j.cell.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duran A., Amanchy R., Linares J.F., Joshi J., Abu-Baker S., Porollo A., Hansen M., Moscat J., Diaz-Meco M.T. p62 is a key regulator of nutrient sensing in the mTORC1 pathway. Mol. Cell. 2011;44:134–146. doi: 10.1016/j.molcel.2011.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tummers B., Green D.R. Caspase-8: regulating life and death. Immunol. Rev. 2017;277:76–89. doi: 10.1111/imr.12541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang S., Okamoto K., Yu C., Sinicrope F.A. p62/sequestosome-1 up-regulation promotes ABT-263-induced caspase-8 aggregation/activation on the autophagosome. J. Biol. Chem. 2013;288:33654–33666. doi: 10.1074/jbc.M113.518134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanchez-Garrido J., Sancho-Shimizu V., Shenoy A.R. Regulated proteolysis of p62/SQSTM1 enables differential control of autophagy and nutrient sensing. Sci. Signal. 2018;11 doi: 10.1126/scisignal.aat6903. [DOI] [PubMed] [Google Scholar]
- 15.Liau P.R., Wu M.S., Lee C.K. Inhibitory effects of scutellaria baicalensis root extract on linoleic acid hydroperoxide-induced lung mitochondrial lipid peroxidation and antioxidant activities. Molecules. 2019;24 doi: 10.3390/molecules24112143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou W., Liu X., Zhang X., Tang J., Li Z., Wang Q., Hu R. Oroxylin A inhibits colitis by inactivating NLRP3 inflammasome. Oncotarget. 2017;8:58903–58917. doi: 10.18632/oncotarget.19440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei L., Dai Y., Zhou Y., He Z., Yao J., Zhao L., Guo Q., Yang L. Oroxylin A activates PKM1/HNF4 alpha to induce hepatoma differentiation and block cancer progression. Cell Death Dis. 2017;8 doi: 10.1038/cddis.2017.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zou M., Lu N., Hu C., Liu W., Sun Y., Wang X., You Q., Gu C., Xi T., Guo Q. Beclin 1-mediated autophagy in hepatocellular carcinoma cells: implication in anticancer efficiency of oroxylin A via inhibition of mTOR signaling. Cell. Signal. 2012;24:1722–1732. doi: 10.1016/j.cellsig.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 19.Tsuboyama K., Koyama-Honda I., Sakamaki Y., Koike M., Morishita H., Mizushima N. The ATG conjugation systems are important for degradation of the inner autophagosomal membrane. Science. 2016;354:1036–1041. doi: 10.1126/science.aaf6136. [DOI] [PubMed] [Google Scholar]
- 20.Jin Z.Y., Li Y., Pitti R., Lawrence D., Pham V.C., Lill J.R., Ashkenazi A. Cullin3-Based polyubiquitination and p62-dependent aggregation of caspase-8 mediate extrinsic apoptosis signaling. Cell. 2009;137:721–735. doi: 10.1016/j.cell.2009.03.015. [DOI] [PubMed] [Google Scholar]
- 21.Jin H., Seo G.S., Lee S.H. Isoliquiritigenin-mediated p62/SQSTM1 induction regulates apoptotic potential through attenuation of caspase-8 activation in colorectal cancer cells. Eur. J. Pharmacol. 2018;841:90–97. doi: 10.1016/j.ejphar.2018.10.015. [DOI] [PubMed] [Google Scholar]
- 22.Martens S. A division of labor in mTORC1 signaling and autophagy. Sci. Signal. 2018;11 doi: 10.1126/scisignal.aav3530. [DOI] [PubMed] [Google Scholar]
- 23.Sanz L., Diaz-Meco M.T., Nakano H., Moscat J. The atypical PKC-interacting protein p62 channels NF-kappaB activation by the IL-1-TRAF6 pathway. EMBO J. 2000;19:1576–1586. doi: 10.1093/emboj/19.7.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qiao C., Wei L., Dai Q., Zhou Y., Yin Q., Li Z., Xiao Y., Guo Q., Lu N. UCP2-related mitochondrial pathway participates in oroxylin A-induced apoptosis in human colon cancer cells. J. Cell. Physiol. 2015;230:1054–1063. doi: 10.1002/jcp.24833. [DOI] [PubMed] [Google Scholar]
- 25.Zhu L., Zhao L., Wang H., Wang Y., Pan D., Yao J., Li Z., Wu G., Guo Q. Oroxylin A reverses P-glycoprotein-mediated multidrug resistance of MCF7/ADR cells by G2/M arrest. Toxicol. Lett. 2013;219:107–115. doi: 10.1016/j.toxlet.2013.01.019. [DOI] [PubMed] [Google Scholar]
- 26.Liu W., Mu R., Nie F.F., Yang Y., Wang J., Dai Q.S., Lu N., Qi Q., Rong J.J., Hu R., Wang X.T., You Q.D., Guo Q.L. MAC-related mitochondrial pathway in oroxylin-A-induced apoptosis in human hepatocellular carcinoma HepG2 cells. Cancer Lett. 2009;284:198–207. doi: 10.1016/j.canlet.2009.04.021. [DOI] [PubMed] [Google Scholar]
- 27.Chen W., Zhang Z., Yao Z., Wang L., Zhang F., Shao J., Chen A., Zheng S. Activation of autophagy is required for Oroxylin A to alleviate carbon tetrachloride-induced liver fibrosis and hepatic stellate cell activation. Int. Immunopharmacol. 2018;56:148–155. doi: 10.1016/j.intimp.2018.01.029. [DOI] [PubMed] [Google Scholar]
- 28.Jin Z., Li Y., Pitti R., Lawrence D., Pham V.C., Lill J.R., Ashkenazi A. Cullin3-based polyubiquitination and p62-dependent aggregation of caspase-8 mediate extrinsic apoptosis signaling. Cell. 2009;137:721–735. doi: 10.1016/j.cell.2009.03.015. [DOI] [PubMed] [Google Scholar]
- 29.Kageyama S., Saito T., Obata M., Koide R.H., Ichimura Y., Komatsu M. Negative regulation of the Keap1-Nrf2 pathway by a p62/Sqstm1 splicing variant. Mol. Cell. Biol. 2018;38 doi: 10.1128/MCB.00642-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Foster A., Scott D., Layfield R., Rea S.L. An FTLD-associated SQSTM1 variant impacts Nrf2 and NF-kappaB signalling and is associated with reduced phosphorylation of p62. Mol. Cell. Neurosci. 2019;98:32–45. doi: 10.1016/j.mcn.2019.04.001. [DOI] [PubMed] [Google Scholar]
- 31.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 32.Bjorkoy G., Lamark T., Pankiv S., Overvatn A., Brech A., Johansen T. Monitoring autophagic degradation of p62/SQSTM1. Methods Enzymol. 2009;452:181–197. doi: 10.1016/S0076-6879(08)03612-4. [DOI] [PubMed] [Google Scholar]
- 33.Norman J.M., Cohen G.M., Bampton E.T. The in vitro cleavage of the hAtg proteins by cell death proteases. Autophagy. 2010;6:1042–1056. doi: 10.4161/auto.6.8.13337. [DOI] [PubMed] [Google Scholar]
- 34.Moscat J., Diaz-Meco M.T. p62: a versatile multitasker takes on cancer. Trends Biochem. Sci. 2012;37:230–236. doi: 10.1016/j.tibs.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H.B., Chen F. Isolation and purification of baicalein, wogonin and oroxylin A from the medicinal plant Scutellaria baicalensis by high-speed counter-current chromatography. J. Chromatogr. A. 2005;1074:107–110. doi: 10.1016/j.chroma.2005.03.088. [DOI] [PubMed] [Google Scholar]
- 36.Zhao Y., Guo Q., Zhu Q., Tan R., Bai D., Bu X., Lin B., Zhao K., Pan C., Chen H., Lu N. Flavonoid VI-16 protects against DSS-induced colitis by inhibiting Txnip-dependent NLRP3 inflammasome activation in macrophages via reducing oxidative stress. Mucosal Immunol. 2019;12:1150–1163. doi: 10.1038/s41385-019-0177-x. [DOI] [PubMed] [Google Scholar]
- 37.Zhao Y., Sun Y., Ding Y., Wang X., Zhou Y., Li W., Huang S., Li Z., Kong L., Guo Q., Lu N. GL-V9, a new synthetic flavonoid derivative, ameliorates DSS-induced colitis against oxidative stress by up-regulating Trx-1 expression via activation of AMPK/FOXO3a pathway. Oncotarget. 2015;6:26291–26307. doi: 10.18632/oncotarget.4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.