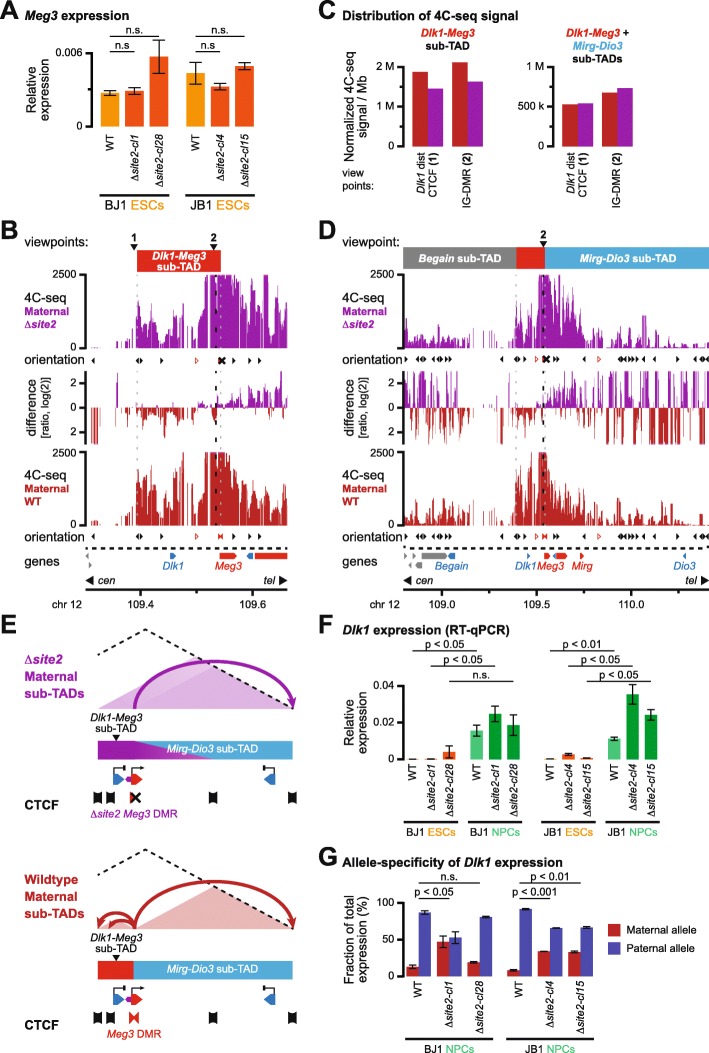
Fig. 4.
Allelic CTCF binding at the Meg3 DMR is essential for correct sub-TAD organization. a Meg3 expression in hybrid ESCs is maintained upon deletion of CTCF binding site 2 in the Meg3 DMR, as determined by qRT-PCR. Error bars indicate standard error of the mean (SEM), with significance of difference determined using a two-sided unpaired t test (n = 2). n.s., not significant. b 4C-seq signal for the IG-DMR (viewpoint 2) on the maternal alleles in hybrid ESCs with a deleted CTCF site 2 in the Meg3 DMR (purple) or their WT counterparts (red) in a 300-kb region around the Dlk1-Dio3 DMRs. The ratio of interactions is provided in between. The orientation of CTCF sites is indicated below each panel, with the X indicating the deleted CTCF site. Viewpoints and the maternal Dlk1-Dio3 sub-TAD are indicated above. c Distribution of 4C-seq signal for indicated viewpoints in the Dlk1-Meg3 sub-TAD (left) and the combined Dlk1-Meg3 and Mirg-Dio3 sub-TADs (right). d 4C-seq signal for the IG-DMR viewpoint across the entire Dlk1-Dio3 TAD. The position of the sub-TADs (red box: Dlk1-Meg3 sub-TAD) is indicated above. e Schematic depiction of the maternal Dlk1-Meg3 and Mirg-Dio3 sub-TAD reorganization upon deletion of CTCF binding site 2 in the Meg3 DMR. CTCF clusters (banners), allele specifically expressed genes in normal cells (triangles), and reported regulatory elements (hexagons) are indicated. f Dlk1 expression levels in hybrid ESCs and in vitro differentiated NPCs with a deleted CTCF site 2 in the Meg3 DMR and their WT counterparts, as determined by qRT-PCR. Error bars indicate SEM, with significance of difference determined using a two-sided unpaired t test (n = 2). g Allelic Dlk1 expression becomes relaxed in hybrid NPCs carrying a deletion in CTCF binding site 2 in the Meg3 DMR, as determined by qRT-PCR. Error bars indicate SEM, with significance of difference between the maternal alleles determined using a one-sided unpaired t test (n = 2)