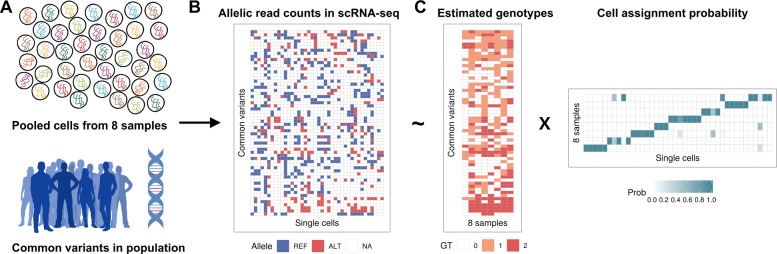
Fig. 1.
Illustration of Vireo for demultiplexing multi-sample scRNA-seq studies without reference genotype data. a, b The inference is based on genotyped common polymorphic variants in each cell, defined based on a standard reference of common human variants. b, c The resulting sparse read count matrices of alternative and reference alleles (displayed as compound matrix for simplicity; NA in white denotes no observed reads) are then decomposed into a matrix of estimated genotypes for each input sample and a probabilistic cell assignment matrix