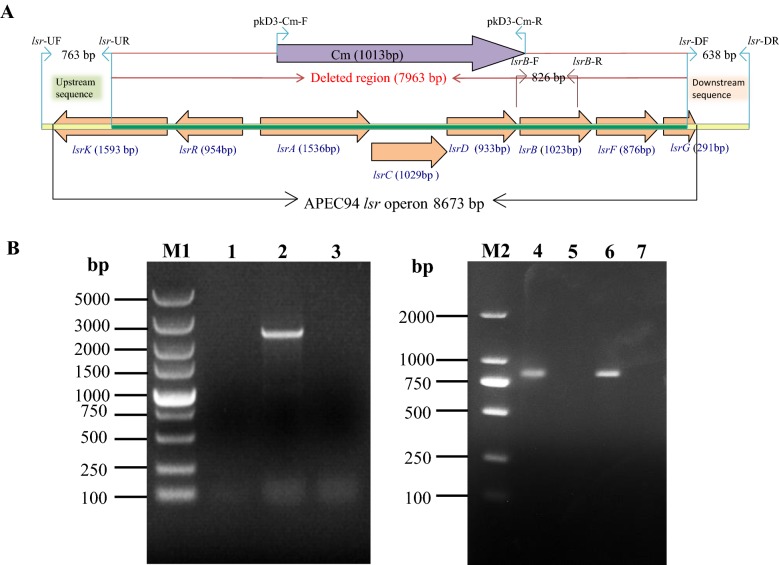
Figure 1.
PCR analysis of the lsr mutant strain APEC94Δlsr(Cm) and lsr complementary strain lsr-APEC94Δlsr. A Schematic chart of the strategy for producing the lsr operon deletion mutant. The lsr operon was deleted by replacing the partial gene sequence with a chloramphenicol resistance cassette. The primers used for confirmation of the lsr deletion are also indicated. B Identification of the lsr mutant strain APEC94Δlsr(Cm) and lsr complementary strain lsr-APEC94Δlsr. M1: 5000 DNA marker; M2: 2000 DNA marker; Lane 1: The wild-type strain APEC94 showed no PCR product using primers lsr-UF/lsr-DR because the amplification time was limited beyond the amplification efficiency of DNA polymerase; Lane 2: The mutant strain APEC94Δlsr(Cm) showed a 2414-bp PCR product using primers lsr-UF/lsr-DR, which amplified upstream and downstream sequences and the chloramphenicol resistance cassette; Lane 3: Negative control; Lane 4: The wild-type strain APEC94 showed a 826-bp PCR product using primers lsrB-F/lsrB-R; Lane 5: The mutant strain APEC94Δlsr(Cm) showed no PCR product using primers lsrB-F/lsrB-R; Lane 6: The lsr complementary strain lsr-APEC94Δlsr showed a 826-bp PCR product using primers lsrB-F/lsrB-R; Lane 7: Negative control.