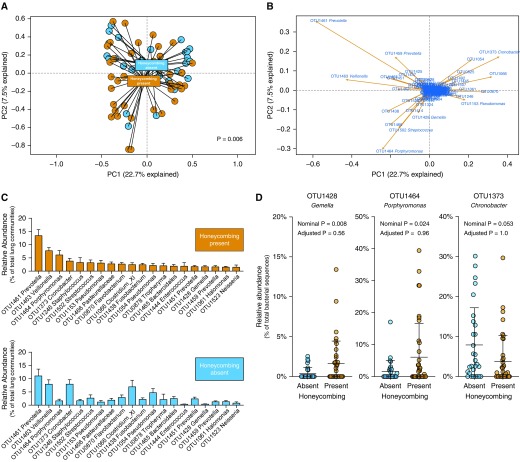
Figure 1.
In patients with idiopathic pulmonary fibrosis, radiographic honeycombing is associated with changes in lung microbiota. Sixty-eight patients with idiopathic pulmonary fibrosis were stratified by the presence or absence of honeycombing on computed tomography scan. Lung microbiota were analyzed using amplicon sequencing of the 16S rRNA gene. Operational taxonomic unit (OTU) data in the principal component (PC) analysis were Hellinger transformed to allow for better behaviors with longer species gradients. (A) Ordination of specimens by community composition (principal component analysis) revealed clustering by honeycombing status, although considerable overlap was observed. Overall community composition was significantly distinct across groups (P = 0.006, mvabund). (B) BiPlot analysis of this ordination identified candidate bacterial taxa responsible for driving the separation of honeycombing specimens (e.g., OTU1428 Gemella and OTU1464 Porphyromonas). (C) Rank abundance analysis revealed taxonomic differences between groups. The 20 most abundant taxa in honeycombing-present specimens are shown in decreasing order of mean relative abundance (mean ± SEM). (D) Specific bacterial taxa were then compared across groups using direct comparison of mean relative abundance. Although select taxa were nominally significant when compared directly (e.g., OTU1428 Gemella and OTU1373 Chronobacter), no specific taxa were significant when adjusted for multiple comparisons by mvabund. Significance was determined via mvabund, and the nominal P values presented were obtained before adjustment for multiple comparisons (A and D).